Projects
Project A

We are interested in the question of how the ubiquitin system controls the function of RNA Polymerase II at core promoters.
The reason for our interest is that the control of RNA Polymerase function by MYC proteins is a central process that drives the genesis of many tumors. Work by many laboratories has shown that MYC oncoproteins are global regulators of transcription and we have shown that degradation of MYC by the ubiquitin system is required for transcriptional elongation (Jaenicke et al., 2016).
We now aim to understand the underlying mechanisms and we work on several strategies to alter MYC degradation in order to control MYC-driven transcriptional regulation and tumorigenesis (Peter et al., 2014).
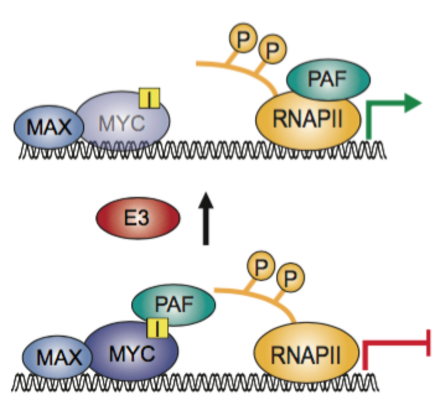
Figure Legend: Degradation of MYC drives transcriptional elongation. PAF1 is a complex that is needed for elongation by RNA Polymerase. At core promoters, it can bind MYC and then represses transcription. Degradation of MYC releases PAF1 to travel with the RNA Polymerase (see Jaenicke et al.,2016).
Jaenicke, L.A., von Eyss, B., Carstensen, A., Wolf, E., Xu, W., Greifenberg, A.K., Geyer, M., Eilers, M., and Popov, N. (2016). Ubiquitin-Dependent Turnover of MYC Antagonizes MYC/PAF1C Complex Accumulation to Drive Transcriptional Elongation. Mol Cell 61, 54-67.
Peter, S., Bultinck, J., Myant, K., Jaenicke, L.A., Walz, S., Muller, J., Gmachl, M., Treu, M., Boehmelt, G., Ade, C.P., et al. (2014). Tumor cell-specific inhibition of MYC function using small molecule inhibitors of the HUWE1 ubiquitin ligase. EMBO molecular medicine 6, 1525-1541.
Project B

The ubiquitin system controls countless physiological and pathophysiological processes and provides a powerful arsenal for drug discovery.
Due to their high specificity ubiquitin ligases (E3 enzymes) present particularly attractive therapeutic targets. This project aims to elucidate the catalytic and regulatory mechanisms, specificities, and interaction networks of HECT-type ligases that are involved in the regulation of transcription and tumorigenic signaling. Our goal is to identify and exploit new target sites for cancer-therapeutic intervention. We will illuminate the molecular architectures of HECT ligases and the macromolecular assemblies they engage in a multi-disciplinary approach using X-ray crystallography, cryo-EM, and NMR in combination with biochemical and biophysical techniques. Structure-based hypotheses will be tested functionally in vitro and in cells.
For a representative study on the HECT ligase HUWE1 from the Lorenz laboratory, please see Sander et al., eLife, 2017.
Project C

Homepage
We are interested in two main tumour types: lung- and colorectal cancer.
By utilising in vitro and in vivo models, human and mouse, we aim to understand the consequence of loss/mutation of key-components of the ubiquitination machinery during tumour initiation, and ways to counteract increased protein stability as a therapeutic approach, by specifically interrogating the protein class of Deubiquitylases.
Project D

In the project we will be interested in mapping replication to have a closer look where replication/transcription conflicts occur.
From proteomic analysis we know that several players of the ubiquitin machinery are associated with MYCN. Since the ubiquitin system is known to play a role in transcription we want to decipher changes in transcription as well as replication/transcription conflicts that will occur at core promoters when we target the ubiquitin system.
Project E

The transcription factor Myc plays a central role in tumourigenesis but was deemed undruggable as no specific inhibitors exist and due to it being an essential protein. However, recent proof-of-principle studies in mice using a dominant negative allele of Myc demonstrated the dependency of established tumours on Myc function and showed that mice tolerated Myc inhibition to a degree that allowed tumour regression.
In line with these observations my group found Myc to regulate distinct sets of genes at low, physiological and high, oncogenic levels, because promoters differ in their affinity for Myc. This notion implies the compelling possibility to specifically target the oncogenic functions of Myc by partial inhibition. We aim to develop strategies to interfere with the oncogenic functions of Myc by developing a novel class of drugs that reduce Myc’s concentrations by utilizing the cellular protein degradation machinery.