More: Structures of two Key Intermediates of the snRNP assembly

Two Crystal Sructures Shed Light onto the Mechanism of snRNP Assembly
Macromolecular complexes facilitate many important tasks in the cell. These complexes are constantly assembled with high accuracy from individual subunits amidst high concentrations of other biological macromolecules. The assembly of macromolecular complexes in vivo is often aided by assembly chaperones, a group of proteins involved in the formation of kinetically trapped intermediates. To proceed in the respective assembly pathway, additional factors are needed to release the imposed kinetic trap. From the pathway of the assisted assembly of U snRNP particles, we have chosen two important and representative intermediates for further structural investigations:
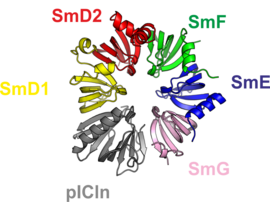
1. A complex consisting of five Sm proteins and the assembly chaperone pICln. This complex is called 6S assembly intermediate, the number referring to the particle’s size as a measure of sedimentation speed. The 6S complex constitutes a major reservoir of Sm proteins in the cell.
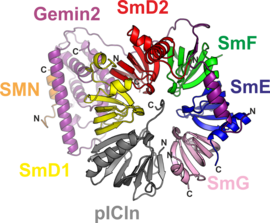
2. An 8S complex that represents an intermediate of the ‘chaperone release’ reaction. The factor needed for release of the chaperone pICln is the SMN complex. This large molecular entity acts as an enzyme during the assembly process and is represented in this engineered complex variant by a minimal set of only two proteins, SMN and Gemin2. We were able to elucidate the atomic structures of both protein complexes with the help of genetic engineering techniques to tailor complex variants that have a high propensity to crystallize.
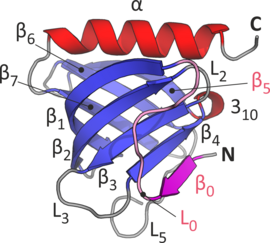
The crystal structure of the 6S complex shows that the five Sm proteins and the assembly chaperone integrate into a doughnut-like structure (Fig. 1). Despite the fact that the chaperone pICln features a folding topology disparate from that of the Sm proteins, it integrates into the torus’ shape in a similar way. This molecular mimicry is enabled by an unanticipated and surprising incorporation of the chaperone’s N-terminus (Fig. 2) into the continuous beta-sheet that spans the whole structure.
The structure of the 8S complex elucidates how the SMN complex binds to the oligohexameric Sm/chaperone ring (Fig. 3). The interactions involved are centered about the tip of a protruding helix of the Gemin2 fold.
Video: We demonstrate the action of the SMN complex by a technique called Normal Mode Analysis.
Normal Mode Analysis of the SMN complex