Progression through the cell cycle involves changes in transcriptional activity, which ensures that proteins required in different cell-cycle phases are produced at the correct time. When cell cycle progression is disrupted, this can lead to a number of diseases such as cancer. The interest of our laboratory is to elucidate pathways involved in transcriptional regulation of cell cycle genes by using a combination of biochemical, cellular and genetic approaches. Our research focuses on the mammalian DREAM and MMB protein complexes, their target genes and their crosstalk with other signaling pathways such as the Hippo-YAP pathway
MuvB complexes: DREAM and MMB
MuvB complexes regulate gene expression through the cell cycle. Dependent on its interactions with different binding partners, MuvB can either repress or activate transcription. When the five protein MuvB core interacts with the p130 retinoblastoma protein paralog, and with E2F4, it forms the DREAM complex, which represses a large number of cell cycle genes in non-dividing cells. Upon cell cycle entry, p130 and E2F4 dissociate from the complex and the MuvB core associates with the transcription factor B-MYB. This new MMB complex plays a key role in activation of a set of genes required for mitosis and cytokinesis, such as Plk1, Cyclin B1 and Cdc20.
Crosstalk between the Hippo YAP pathway and MMB
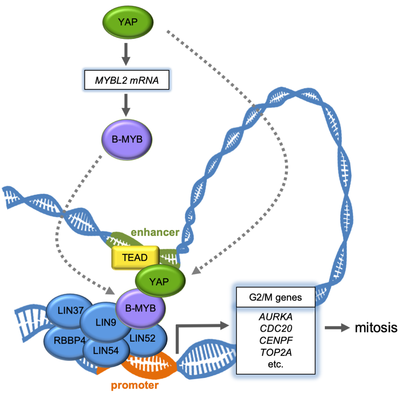
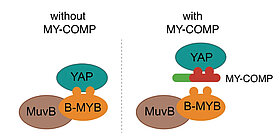
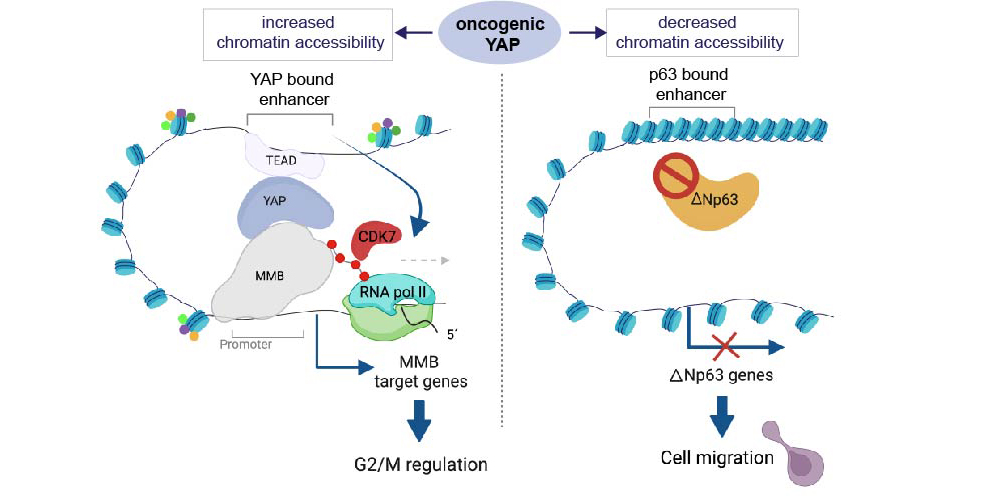
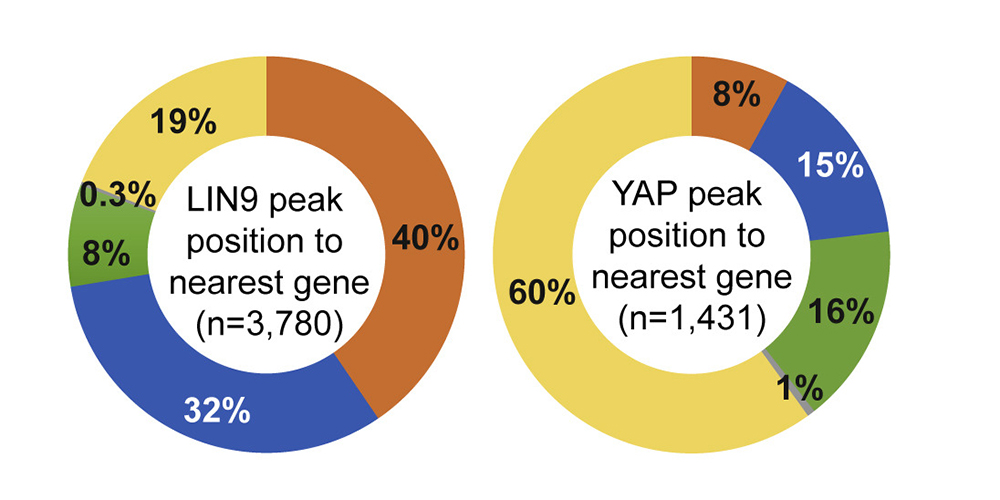
The Hippo pathway and its downstream effectors YAP/TAZ play key roles in the control of cell proliferation, organ size and tumorigenesis. We recently observed that the transcriptional coactivator YAP and MMB co-regulate an overlapping set of late cell cycle genes and that the ability of YAP to activate mitotic genes and stimulate cell cycle progression requires its interaction with the MMB complex. YAP binds to distal enhancers and interacts with MMB regulated promoters through chromatin looping. These findings indicate that two major cancer-relevant signaling pathways are linked.
We use state of the art methods including CRSIPR/Cas9, ChIPseq, RNAseq and ATACseq to investigate gene regulation by YAP, DREAM and MMB.