The three-dimensional architecture of genomes determines the majority of their functions, from the control of gene expression to the correct segregation of chromosomes during mitotic and meiotic cell divisions. The main goal of our research is to reveal the action of molecular machines that organize eukaryotic chromosomes.
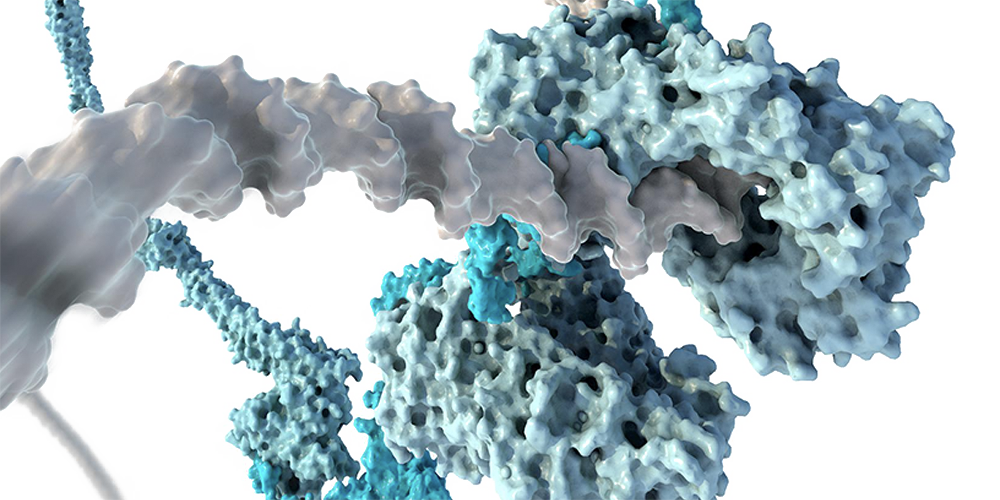
Our work focuses on two members of the Structural Maintenance of Chromosomes (SMC) family of protein complexes: cohesin and condensin. We have shown that condensin is a molecular motor that uses the energy of adenosine triphosphate (ATP) hydrolysis to move along the DNA double helix and thereby fold it into large loop structures. This loop-extrusion acticity presumably forms the basis for structuring chromatin fibers into rod-shaped mitotic chromosomes.
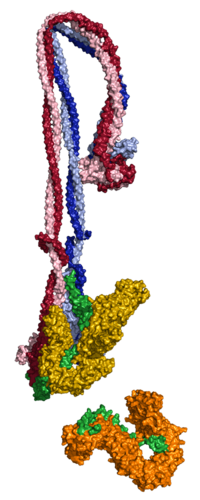
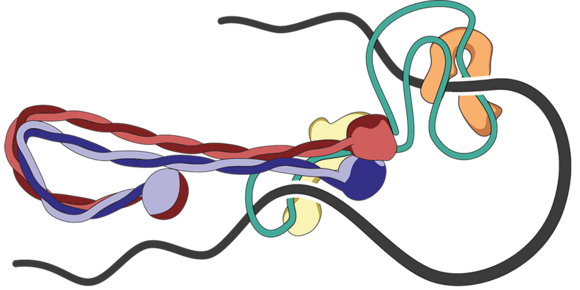
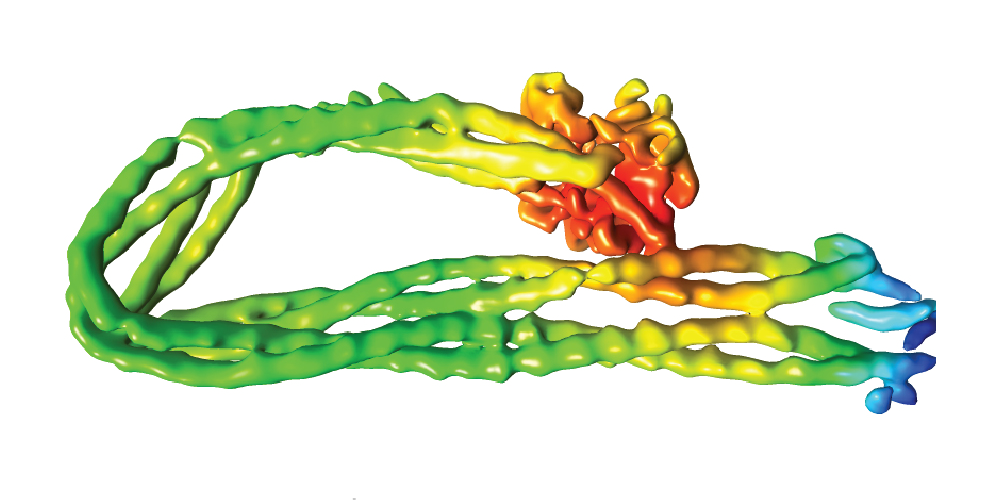
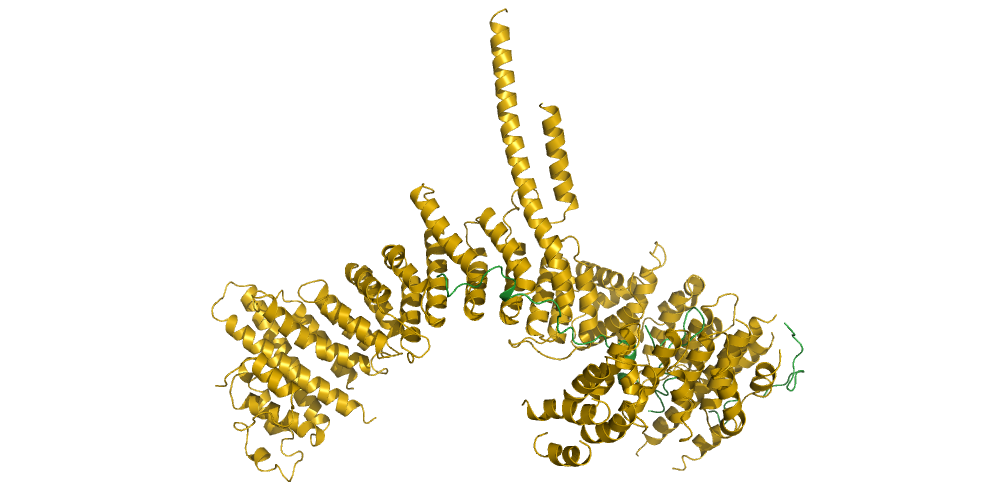
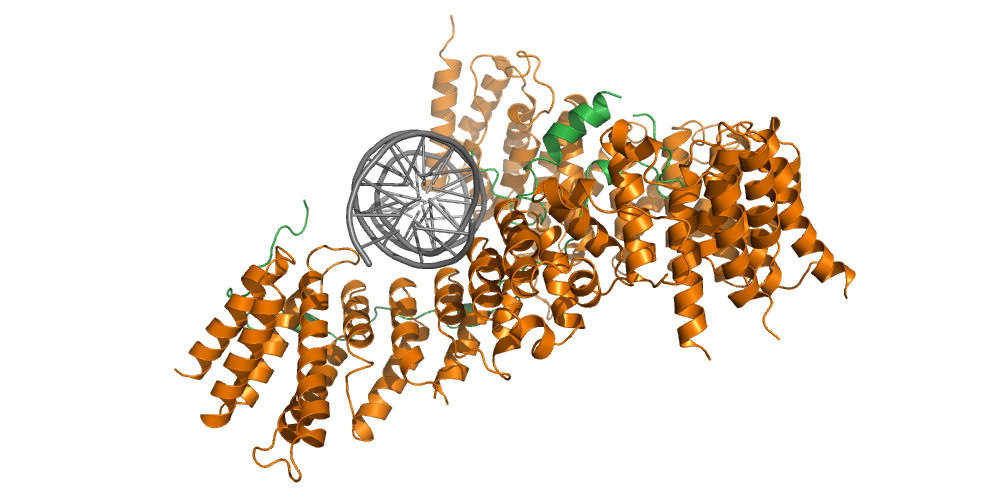
In our group, we combine cutting-edge methods from biochemistry, molecular cell biology, biophysics and structural biology into highly interdisciplinary approaches to study the mechanisms of SMC protein complexes from the atomic to the cellular scale. Using a combination of x-ray protein crystallography and cryo electron-microscopy, we have solved the structure of the entire condensin complex at near-atomic resolution at different states of its ATPase reaction cycle. Mapping of the path of DNA through the complex based on single-molecule microscopy and bulk biochemistry techniques has provided us with a working model that explains how condensin can take uniquely large steps in a single direction while consuming a minumum of energy.
We are currently testing our working model in purified reconstitution systems, in cultured human cells and entire model organisms. Insights into the action of SMC complexes will reveal how they control all aspects of genome biololgy and how their function is affected in various human conditions.