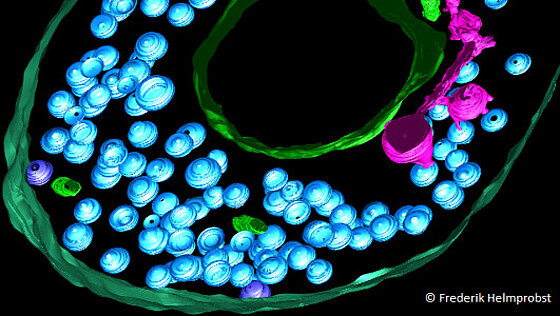
Synaptic Architecture (Stigloher)
Research: Synaptic architecture at the nano-scale
Neuronal synapses are highly efficient and complex cellular signaling machineries that achieve remarkable precision in signal transmission for a prolonged period of time, in some cases throughout the lifetime of an animal. The importance of synaptic efficiency is mirrored by many neural diseases but in particular by synaptopathies where synaptic organization and function is disrupted. In order to provide reliable signaling synaptic vesicles have to be retained close to the presynaptic active zone, the domain where vesicles are docked and fuse with the membrane after Ca influx in a nano-domain through voltage gated channels.
How are synaptic vesicles kept coherently close to the active zone to maintain efficient signaling? To shed light onto this question our team focuses on the cellular architecture using a combination of genetic tools and imaging techniques. In particular we apply electron tomography as ultra high 3D resolution method to dissect components and function of synaptic architecture. We use a synergistic combination of two highly tractable models where they are most appropriate: The C. elegans neuromuscular junctions for efficient candidate identification and manipulation and the neuromuscular junctions of the zebrafish larva as vertebrate model to test for evolutionary conservation of function.
-
(2024) “Interaction of human keratinocytes and nerve fiber terminals at the neuro-cutaneous unit”, eLife, 13, e77761, available: https://doi.org/10.7554/eLife.77761.
- [ URL ]
-
(2024) “Degradation of hexosylceramides is required for timely corpse clearance via formation of cargo-containing phagolysosomal vesicles”, European Journal of Cell Biology, 103(2), 151411, available: https://doi.org/https://doi.org/10.1016/j.ejcb.2024.151411.
- [ URL ]
-
(2024) “Chapter Seven - Array tomography of in vivo labeled synaptic receptors”, in Müller-Reichert, T. and Verkade, P., eds., Correlative Light and Electron Microscopy V, Methods in Cell Biology, Academic Press, 139–174, available: https://doi.org/https://doi.org/10.1016/bs.mcb.2024.02.029.
- [ URL ]
-
(2023) “A BORC-dependent molecular pathway for vesiculation of cell corpse phagolysosomes”, Current Biology, 33(4), 607–621.e7, available: https://doi.org/https://doi.org/10.1016/j.cub.2022.12.041.
- [ URL ]
-
(2021) “Structural Analysis of the Caenorhabditis elegans Dauer Larval Anterior Sensilla by Focused Ion Beam-Scanning Electron Microscopy”, Frontiers in Neuroanatomy, 15, 80, available: https://doi.org/10.3389/fnana.2021.732520.
- [ URL ]
-
(2021) “Click-correlative light and electron microscopy (click-AT-CLEM) for imaging and tracking azido-functionalized sphingolipids in bacteria”, Scientific Reports, 11(1), 4300-, available: https://doi.org/10.1038/s41598-021-83813-w.
- [ URL ]
-
(2020) “Overexpression of an {ALS}-associated {FUS} mutation in C. elegans disrupts {NMJ} morphology and leads to defective neuromuscular transmission”, Biology Open, 9(12), bio055129, available: https://doi.org/10.1242/bio.055129.
- [ URL ]
-
(2020) “Primary and secondary motoneurons use different calcium channel types to control escape and swimming behaviors in zebrafish”, Proceedings of the National Academy of Sciences, 117(42), 26429–26437, available: https://doi.org/10.1073/pnas.2015866117.
- [ URL ]
-
(2020) “Advancing Array Tomography to Study the Fine Ultrastructure of Identified Neurons in Zebrafish (Danio rerio)”, Springer Protocols, Neuromethods(155), 59–78, available: https://link.springer.com/protocol/10.1007%2F978-1-0716-0691-9_4.
- [ URL ]
-
(2018) “Automated classification of synaptic vesicles in electron tomograms of C. elegans using machine learning”, PLOS ONE, 13(10), 1–22, available: https://doi.org/10.1371/journal.pone.0205348.
- [ URL ]
-
(2018) “CRELD1 is an evolutionarily-conserved maturational enhancer of ionotropic acetylcholine receptors”, eLife, 7(e39649), available: https://doi.org/10.7554/eLife.39649.
- [ URL ]
-
(2017) “Expression of sept3, sept5a and sept5b in the Developing and Adult Nervous System of the Zebrafish (Danio rerio)”, Frontiers in Neuroanatomy, 11, available: https://doi.org/10.3389/fnana.2017.00006.
- [ URL ]
-
(2017) “FIJI Macro 3D ART VeSElecT: 3D Automated Reconstruction Tool for Vesicle Structures of Electron Tomograms”, PLOS Computational Biology, 13(1), 1–21, available: https://doi.org/10.1371/journal.pcbi.1005317.
- [ URL ]
-
(2017) “sept8a and sept8b mRNA expression in the developing and adult zebrafish”, Gene Expression Patterns, 25-26, 8–21, available: https://doi.org/https://doi.org/10.1016/j.gep.2017.04.002.
- [ URL ]
-
(2017) “Chapter 2 - 3D subcellular localization with superresolution array tomography on ultrathin sections of various species”, in Müller-Reichert, T. and Verkade, P., eds., Correlative Light and Electron Microscopy III, Methods in Cell Biology, Academic Press, 21–47, available: https://doi.org/https://doi.org/10.1016/bs.mcb.2017.03.004.
- [ URL ]
-
(2017) “Chapter 4 - Minimal resin embedding of multicellular specimens for targeted FIB-SEM imaging”, in Müller-Reichert, T. and Verkade, P., eds., Correlative Light and Electron Microscopy III, Methods in Cell Biology, Academic Press, 69–83, available: https://doi.org/https://doi.org/10.1016/bs.mcb.2017.03.005.
- [ URL ]
-
(2016) “Filling the gap: adding super-resolution to array tomography for correlated ultrastructural and molecular identification of electrical synapses at the C. elegans connectome”, Neurophotonics, 3(4), 041802, available: https://doi.org/10.1117/1.nph.3.4.041802.
- [ URL ]
-
(2015) “Presynaptic architecture of the larval zebrafish neuromuscular junction”, Journal of Comparative Neurology, 523(13), 1984–1997, available: https://doi.org/10.1002/cne.23775.
- [ URL ]