Sex determination and reproductive development in fish
(Manfed Schartl, Mateus Adolfi, Juliana Ricci, Ruiqi Liu, Irene da Cruz, Kang Du, Petra Fischer)
In fish, mechanisms of sex determination are very diversified: even within groups of closely related species a wide spectrum of different systems can be found. We are studying the molecular mechanisms of sex determination and the genetic organization of sex chromosomes in selected fish species to generate a database that will allow a better understanding of the evolutionary processes generating such apparent diversity.
Evolution of sex determination and sex chromosomes
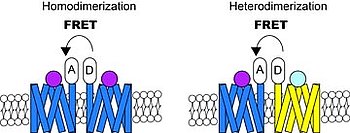
Poeciliids are a suitable animal model to study the evolution of sex determination and sex chromosomes. Species with either male (XX/XY) or female (WZ/ZZ) heterogamety exist within this group and in some cases even three types of sex chromosomes (W, Y, X) have been reported, e.g., in the platyfish (Xiphophorus maculatus).
Sex-linked inheritance of traits has been demonstrated; for instance, many of the highly polymorphic color patterns of the guppy (Poecilia reticulata) and platyfish are linked to the respective male sex-determination locus on the Y chromosome. Nevertheless, the presence of large amounts of repeats, caused by the suppression of recombination around the sex-determining gene, makes it difficult to identify the genes located in these regions.
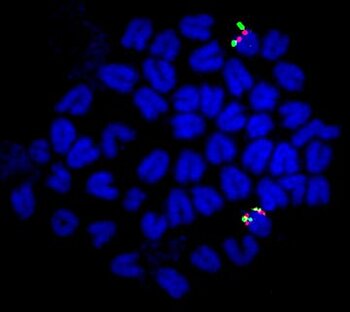
In order to characterize the sex-determining region in the guppy, platyfish, and other poeciliids, we utilize Next Generation Sequencing (NGS) (whole genome sequencing, RNA-seq, and RAD-seq) as well as cytogenetic and molecular biology techniques, for instance, FISH, cloning of candidate genes, in situ hybridization and BAC and fosmid libraries. Identification of the genes in these regions will help to understand how the high degree of color polymorphism is generated and maintained in these species and will reveal the evolutionary relationships of these sex chromosomes.
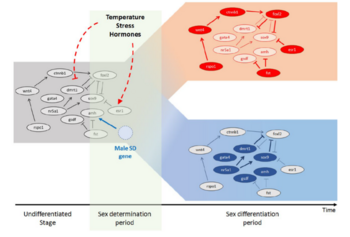
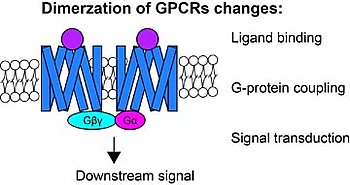
Mechanisms of sex determination and evolution of the sex-regulatory network
The diversity of sex determination mechanisms is particularly evident in species of the genus Oryzias, which includes the medaka, O. latipes. In this fish the master male sex determination gene (dmrt1bY) is known and it is present in the sister species O. curvinotus but absent in other closely related species. However, high diversity of sex-determining genes is observed in several groups of vertebrate species, especially in fish. Identification of different sex-determining genes in vertebrates demonstrated that some genes recurrently became the sex-determining trigger, for instance the members of the Tgf-β signaling pathway. Despite the huge diversity of upstream and downstream factors and networks involved in the process of sex determination, some phenomena still appear to be highly conserved throughout evolution. A common feature found in most studied vertebrates is an early morphological difference between male and female that becomes apparent on the germ cell level. The germ cells start to proliferate and enter meiosis first in female compared to male. Thus, main questions of current interest in our group are: How similar are the downstream gene networks that respond to these triggers and actually how do they work? Did the diversity of sex determination mechanism converge to conserved key effectors, like germ cells proliferation and meiosis entry? We use techniques like gene expression by qRT-PCR and in situ hybridization, transgenic reporter lines, genome editing by CRISPR/Cas9, immunohistochemistry and confocal microscope imaging for medaka. We also study expression of sexual development genes in different fish species (e.g. arapaima, sterlet, guppy, platyfish and swordtails) and compare to our medaka model to understand the evolution of the sex-regulatory network in vertebrates.
Molecular dynamics of Mc4r in the Xiphophorus fish reproduction
Reproduction is a fundamental feature of life. Puberty is an important physiological process in the reproduction, and timing of puberty is a polymorphic trait within species affected by genetic polymorphisms.
Fish of the genus Xiphophorus are a prominant example for a genetic polymorphism in puberty timing and reproductive strategy. As male fish of the genus Xiphophorus, like in many other fish species, cease to grow when they reach puberty, the size of the fish correlates with the timing of the puberty. Wild populations contain naturally “early-matured” small males and “late-matured” large males. Large, late-maturing males have their own territories and display courtship rituals tofemales. Small, early-maturing males, have no territories and perform sneak mating.
In particular in two species, X. nigrensis and X. multilineatus, we showed that the mc4r gene, encoding the melanocortin receptor 4, is the determining factor for timing of puberty onset. This is brought about by different mc4r alleles and copy number variation of these mc4r alleles in late vs early maturing males. The Mc4r signaling regulates energy balance in humans and other mammals and malfunction results in obesity. In our fish model we are interested in the molecular mechanism of Mc4r signalling and how it is modulated by the various alleles and their copy number variation. In order to untangle how Mc4r controls puberty onset on a molecular basis, we use techniques including molecular cloning, transgenic reporter line, luciferase assay, fluorescence resonance energy transfer techniques, confocal or other advanced microscopy imaging.
Evolution of sexual reproduction
(Manfed Schartl, Irene da Cruz)
Sexual reproduction is the most widespread form of reproduction among eukaryotes. The prevalence of sex relies on recombination during meiosis, which is predicted to enhance faster adaptation to changing environments and purging deleterious alleles. In contrast, asexual organisms lack meiotic recombination and are predicted by several evolutionary theories (Muller's ratchet or “Red Queen”) to suffer from low genetic variation. Thus, an asexual species will rapidly become extinct, as their genomes accumulate harmful mutations or are not well adapted to survive environmental changes.
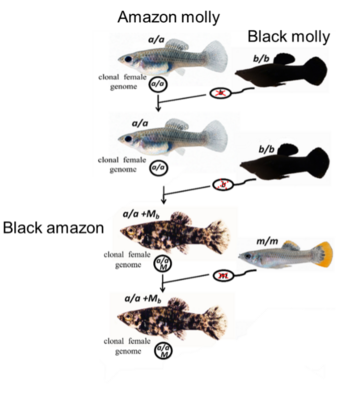
However, the fish Amazon molly (Poecilia formosa), described as the first unisexual vertebrate, has been reproducing asexually for much longer than the mentioned theories had predicted. The reproduction mode of Amazon molly involves the sperm of males from related sexual species as physiological stimulus to the development of the ameiotic egg, but its genetic content is excluded. As result all-female clonal individuals are produced.
In rare cases, parts of the paternal genome from the sperm donor persist as microchromosomes in the developing embryo. Evidence of these infrequent paternal DNA introgression have been observed when Amazon mollies were bred in the laboratory with males of a homogeneously black-colored ornamental Poecilia line, called Black molly. This new Amazon molly pedigree presents an unusual pigmentation phenotype from black-spotted to completely black. The occurrence of the genes for black body pigmentation in the pigmented Amazon molly clone (called Black amazon) most likely derived from the Black molly and was correlated with the presence of stably inherited microchromosomes.
Our goal is to investigate whether such microchromosomes play a role in increasing the genetic diversity and thus adaptability to the long-term success of this all-female asexual species. To achieve this goal, we use bioinformatic approach for analysis of genome-wide sequencing data to identify gene content, genomic organization and expression patterns from an isolated microchromosome derived from Black Amazons.
The all-female clonal Amazon Molly (genome composition a/a) reproduce in the laboratory with males of the ornamental Black molly (b/b) strain through gynogenesis. The sperm only triggers the development of the diploid oocyte (a/a), but its genetic content is excluded (red crosses on the haploid sperm b). The offspring of these laboratory broods generally have wild-type pigmentation, being again clonal females. Unfrequently such offspring presented black-spotted phenotype (genome composition a/a +Mb) most likely derived from the father’s DNA incorporation as microchromosome (Mb) in the egg nucleus. The Amazon mollies black-spotted (a/a +Mb) pedigree continued to reproduce via gynogenesis with unpigmented males (genome constitution m/m), giving rise to clonal female genomes with the pigmented phenotype (a/a +Mb) evidencing that such microchromosomes were inherited over generations.