Prof. Dr. Markus Sauer | |
Telefon: | +49 931 31-88687 |
Super-resolution imaging methods based on single-molecule localization have arrived as valuable tools for studying cellular architecture at the nanoscale [1, 2]. Single-molecule based localization microscopy methods, such as PALM, FPALM, STORM, and dSTORM utilize photoactivatable or -switchable fluorophores, which enable the temporal separation of fluorescence emission. Besides molecular optical photoswitches, localization microscopy requires widefield microscope setups with laser light excitation and sensitive photon detection (e.g., electron multiplying CCD cameras), efficient label strategies, and appropriate software for data analysis.
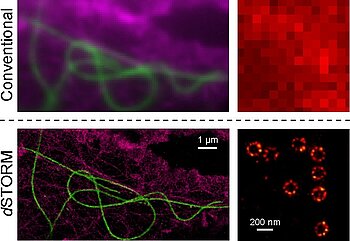
Fig. 1: dSTORM images of cellular nanostructures. Left: Super-resolution imaging of the cytoskeleton; microtubules (green), f-actin (magenta). Right: Super-resolution imaging of the nuclear pores in X. laevis A6 cells.
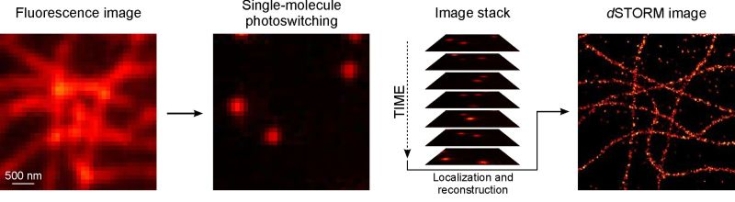
The localization microscopy method dSTORM uses standard fluorescent dyes that can be switched reversibly between a fluorescent and non-fluorescent state upon irradiation with light. Photoswitchable fluorophores linked to molecular probes such as antibodies, toxins, oligonucleotides, or chemical tags are used for selective staining and high resolution imaging of cellular nanostructures [3, 4, 5, 6].
Fig. 2: dSTORM imaging of fixed COS-7 cells. Microtubules were stained with commercially available Alexa Fluor 647 labeled antibodies. Temporal separation of single emitters localized precisely by data modelling enables image reconstruction with superior resolution.
At the beginning of the experiment, the majority of fluorophores is switched to a metastable dark state. Only sparse subsets of individual emitters are switched on stochastically. Single, temporally separated emitters are localized with high spatial precision using Gaussian data modelling. Finally, an artificial super-resolution image is reconstructed from thousands of images and millions of localizations, yielding a resolution better than 20 nm in lateral direction.
Since a single acquisition can easily generate data in the gigabyte range, fast analysis algorithms are crucial to implement dSTORM as a standard laboratory method. Therefore we developed the open-source software rapidSTORM, which enables real-time computation of dSTORM image stacks [7].
1. van de Linde, S., Sauer, M.
How to switch a fluorophore: from undesired blinking to controlled photoswitching.
Chem. Soc. Rev. 2014, 43(4), 1076-1087.
2. van de Linde S, Heilemann M, Sauer M.
Live-cell super-resolution imaging with synthetic fluorophores.
Annu. Rev. Phys. Chem. 2012, 63:519-540.
3. van de Linde S, Aufmkolk S, Franke C, Holm T, Klein T, Loschberger A, Proppert S, Wolter S, Sauer M.
Investigating cellular structures at the nanoscale with organic fluorophores.
Chem. Biol. 2013, 20(1):8-18.
4. van de Linde S, Loschberger A, Klein T, Heidbreder M, Wolter S, Heilemann M, Sauer M.
Direct stochastic optical reconstruction microscopy with standard fluorescent probes.
Nat. Protoc. 2011, 6(7):991-1009.
5. Löschberger, A., van de Linde, S., Dabauvalle, M.-C., Rieger, B., Heilemann, M., Krohne, G., Sauer, M.
Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution.
J. Cell Sci. 2012, 125, 570-575.
6. Ehmann, N., van de Linde et al.
Quantitative super-resolution imaging of Bruchpilot distinguishes active zone states.
Nat. Commun. 2014, 5, 4650.
7. Wolter S, Loschberger A, Holm T, Aufmkolk S, Dabauvalle MC, van de Linde S, Sauer M.
rapidSTORM: accurate, fast open-source software for localization microscopy.
Nat. Methods 2012, 9(11):1040-1041.