sRNA- and asRNA-mediated gene expression control in Neisseria gonorrhoeae
Noncoding (nc) RNAs are well-established as post-transcriptional regulators of bacterial stress responses and bacterial virulence. Bacterial small regulatory ncRNAs (sRNAs) range in size from 50 to 500 nucleotides and are commonly transcribed from intergenic regions of the bacterial genome. These trans-encoded sRNAs show only limited complementarity to their target mRNAs and hybridize at or in close proximity to the ribosome binding site thereby controlling ribosome binding and translation (Fig. 1).
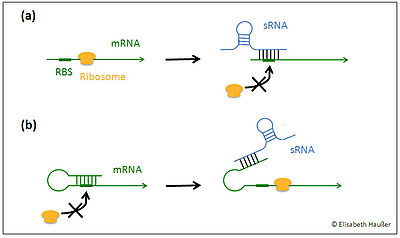
Fig. 1: Common mechanisms of sRNA action in bacteria:
(a) Translation repression: The formation of a mRNA/sRNA complex results in an occlusion of the RBS (ribosome binding site) and thereby prevents translation initiation.
(b) Translation activation: By base pairing with a sequence within the 5'UTR of the target mRNA the sRNA unmasks the RBS and increases target translation.
In addition mRNA stability can be affected by the formation of mRNA-sRNA complexes. The length of cis-encoded antisense RNAs (asRNAs) ranges from less than 100 nucleotides to several kilobases. Their function is less well understood, however, they have been implicated in the modulation of mRNA stability, translation and transcription termination or in transcriptional interference. In-depth analysis of the transcriptome of the sexually transmitted human pathogen Neisseria gonorrhoeae revealed the presence of 253 transcripts which are not derived from a coding sequence (Remmele et al., 2014). 59 of these transcripts mapped to intergenic regions and therefore are likely to function as trans-acting sRNAs. Antisense transcripts for 194 genes were identified representing 16% of all primary transcripts detected in this analysis. The asRNAs are either transcribed from the coding region of the respective gene or represent long untranslated 5’ or 3’ sequences of an mRNA which overlap with the mRNA of a gene which is transcribed in the opposite direction. Furthermore, sRNAs and mRNAs bound by the RNA chaperone Hfq were identified by co-immunoprecipitation of FLAG-tagged Hfq and subsequent deep sequencing. The aim of this project is the functional analysis of selected sRNAs and asRNAs of N. gonorrhoeae. Interestingly, asRNAs were found to be frequently associated with genes which are subject to phase variation via slipped strand mispairing due to the presence of sequence repeats within the coding region. Therefore, a key aspect of the project addresses the role of these asRNAs in the turnover of in-frame and out-of-frame mRNAs.