Microbiota
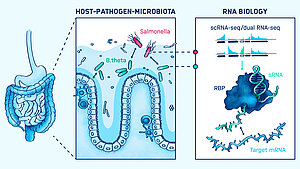
Our intestinal tract offers an attractive environment for both beneficial and pathogenic bacteria. The beneficial bacteria of our microbiota feast on undigested foods and provide numerous health benefits. Enteric pathogens see this environment as an entry point for infection. Both groups influence each other, creating a tripartite interaction with us, the host. Understanding the regulatory processes that decide on the outcome of these encounters represents an emerging research area to combat infectious diseases. While the field has focused on protein-mediated processes, our group investigates the role of RNA-centric mechanisms in controlling microbial interactions in the gut.
Our current work focuses on the transcriptome of human gut commensal Bacteroides thetaiotaomicron in the context of host niche colonization in the human colon. We develop in vitro colonization models and utilize RNA-sequencing technologies, such as Dual RNA-seq and Triple RNA-seq. The group’s overarching goal is to find novel targets for the treatment and prevention of enteric infections based on RNA mechanisms.