ADHESION
Cell adhesion
is more than a handshake between binders. What role does physics play?
Model systems
allow to unequivocally separate physical processes from active biological cell responses.
Microinterferometry
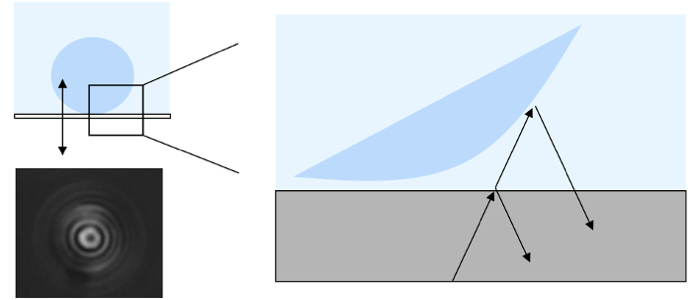
RICM images contain intensity-coded height information.
Cell-cell adhesion
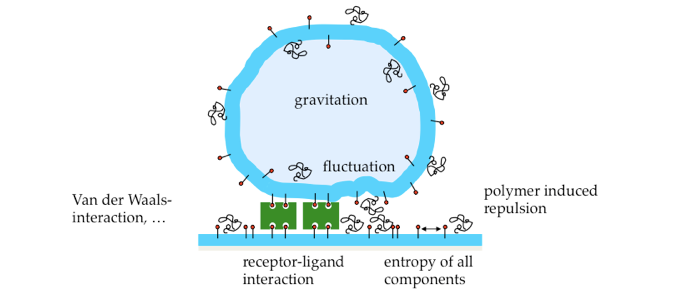
Adhesion of cells to each other is of vital importance for any multicellular organism. It controls how cells connect to form one tissue (e.g. during wound healing) or separate to form different tissues (e.g. during development).
Adhesion is mediated by specific bonds between cell surface molecules. The major cell adhesion molecules responsible for calcium-dependent cell-cell adhesion in vertebrate tissue are cadherins. These are trans-membrane proteins that predominantly bind other cadherins in a homophilic fashion. In mature adhesions, they form supra-molecular structures (so-called adherens junctions), which are connected to the cytoskeleton inside the cell, thus forming a mechano-chemical link between cells. Important physical and biological events occur in between the very first recognition of an adhesive surface and the formation of mature contact structures. Our work aims at a quantitative understanding of the physical processes in the early stages of cell-cell adhesion caused by mobile cell adhesion molecules. Since cell adhesion molecules reside on the cell membrane, the membrane itself is a crucial player.
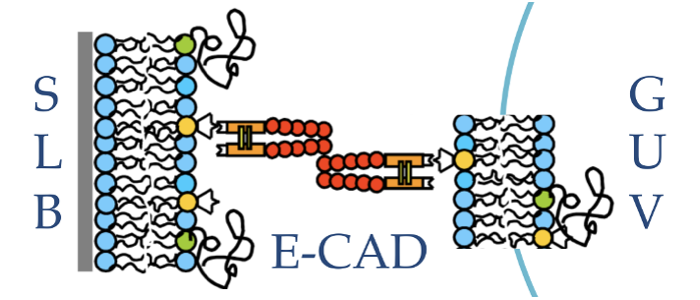
Model system. To unequivocally separate physical processes from active biological cell responses, a biomimetic model system for cell adhesion was developed. It consists of a giant unilamellar vesicle (GUV) adhering via specific ligand-receptor interactions to a solid supported lipid bilayer (SLB) or a micro-patterned surface. Adhesion is mediated by either biotin-neutravidin (an avidin analogue) or the extracellular domains of the cell-cell adhesion molecule E-cadherin.
Physical parameters
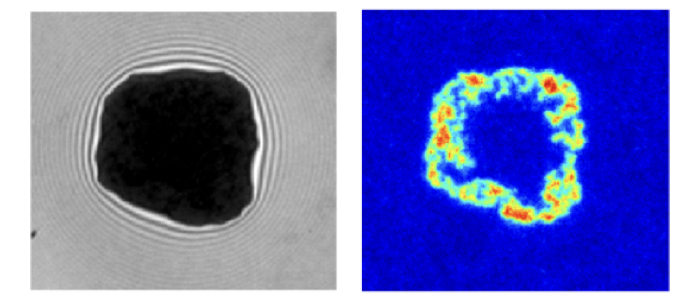
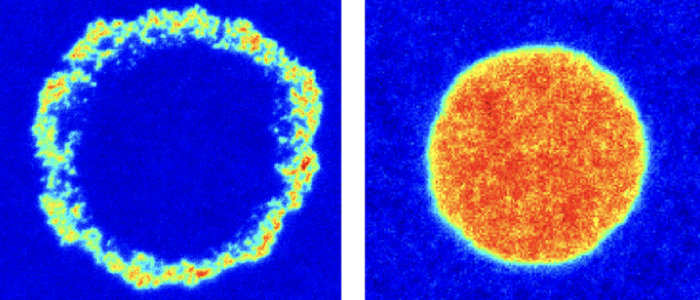
A combination of fluorescence microscopy and interferometry is applied to study the influence of receptor mobility and membrane fluctuations on adhesion dynamics, steady state as well as the geometry of the adhesion cluster in steady state. In the biotin-neutravidin system, the neutravidin receptor accumulates in the vesicle adhesion zone and fills the zone completely resulting in strong adhesion or forms a ring along the periphery resulting in weak adhesion. Surprisingly, the different cluster types and adhesion states form in a purely passive model system using the same intrinsically strong binder.
Using the E-cadherin model system it was found that bending fluctuations of membranes are a source of long-range cis-interactions between cadherins, and regulate their trans-interactions. This regulation relies on physical principles and not on details of protein-protein interactions. These omnipresent fluctuations can thus act as a generic control mechanism in all types of cell adhesion.
Reflection interference contrast microscopy (RICM) is used to reconstruct height profiles of objects from the interference pattern of light being reflected at the objects’ surface. RICM images contain intensity-coded height information. Next to binding heights, membrane fluctuations and on-rates can be determined. Using dynamical RICM nanometric adhesion domains are identified label-free using suppression of membrane fluctuations as a signature for binding.
Selected publications
Fenz SF, Bihr T, Schmidt D, Merkel R, Sengupta K, Seifert U, Smith A.-S. (2017), Membrane fluctuations mediate lateral interaction between cadherin bonds, Nature Physics, doi: 10.1038/NPHYS4138.
Schmidt D, Bihr T, Fenz SF, Merkel R, Seifert U, Sengupta K, Smith AS, (2015), Crowding of receptors induces ring-like adhesions in model membranes, BBA: Molecular Cell Research, doi: 10.1016/j.bbamcr.2015.05.025
Bihr T, Fenz SF, Sackmann E, Merkel R, Seifert U, Sengupta K, Smith AS (2015) Association Rates of Membrane-Coupled Cell Adhesion Molecules. Biophysical Journal Letter, doi: 10.1016/j.bpj.2014.10.033
Monzel C, Fenz S, Giesen M, Merkel R, Sengupta K (2012), Mapping fluctuations in biomembranes adhered to micropatterns, Soft Matter, doi: 10.1039/c2sm07458c
Fenz SF, Sengupta K (2012) Giant Vesicles as Cell Models, Integrative Biology, doi: 10.1039/c2ib00188h
Fenz SF, Smith AS, Merkel R, Sengupta K (2011) Inter-membrane Adhesion Mediated by Mobile Linkers: Effect of Receptor Shortage, Soft Matter, doi: 10.1039/c0sm00550a
Fenz SF, Bihr T, Merkel R, Seifert U, Sengupta K, Smith AS (2011), Switching from ultra-weak to strong adhesion., Advanced Materials, doi: 10.1002/adma.201004097
Smith AS, Fenz SF, Sengupta K (2010), Inferring spatial organization of bonds within adhesion clusters by exploiting fluctuations of soft interfaces, Europhysics Letters, doi: 10.1209/0295-5075/89/28003
Monzel C, Fenz S, Merkel R, Sengupta K (2009), Probing Biomembrane Dynamics by Dual-Wavelength Reflection Interference Contrast Microscopy, ChemPhysChem, doi: 10.1002/cphc.200900645
Fenz SF, Merkel R, Sengupta K (2009) Diffusion and Inter-Membrane Distance: Case Study of Avidin and E-cadherin Mediated Adhesion, Langmuir, doi: 10.1021/la803227s