mRNA export
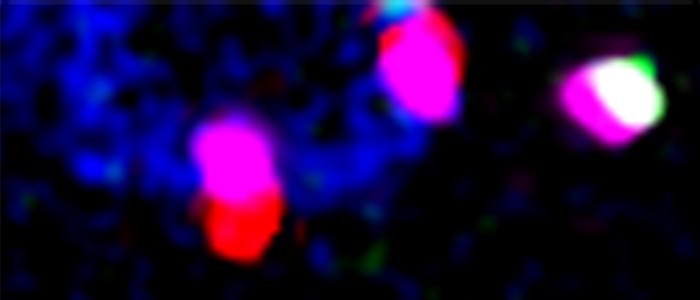
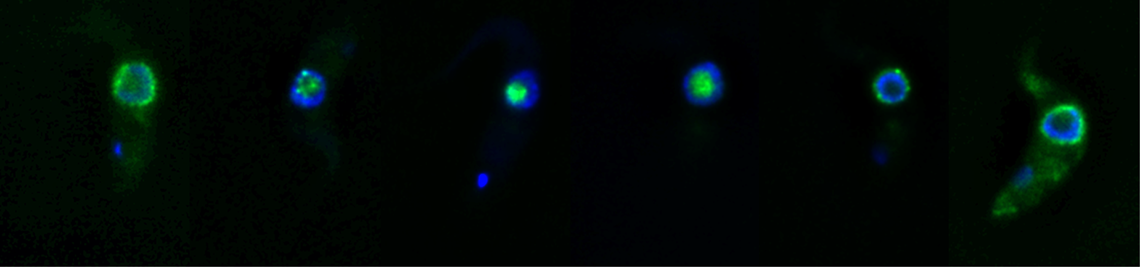
Co-transcriptional nuclear export in trypanosomes, visualised by intramolecular three-colour single molecule FISH. The large reporter mRNA FUTSCH is stained in red at the 5´end, in green at the 3´end and in infrared (pink false colours) throughout the large middle part. Red-pink molecules across the nuclear envelope (bottom) indicate mRNAs in transcription that have started nuclear export.
Co-transcriptional nuclear export
mRNAs are transcribed in the nucleus and are exported to the cytoplasm via the largest protein complex of the cell, the nuclear pore complex. In most organisms, an elaborate quality control system ensures that only fully transcribed and processed mRNAs can enter the cytoplasm, to avoid the production of aberrant proteins. Trypanosomes have unusually symmetric nuclear pores, lack homologues to most of the nuclear export control proteins and non-conventionally transcribe mRNAs polycistronically followed by processing via trans-splicing. Perhaps this is the reason for the novel and highly unusual way of co-transcriptional nuclear export that we recently discovered in these parasites: trypanosomes can start (but not finish) nuclear export of mRNAs have not been fully transcribed or processed. Inhibition of trans-splicing causes the formation of trypanosome-unique RNA granules at the cytoplasmic site of the nuclear pores (NPGs, nuclear pore granules). NPGs contain most RNA binding proteins (except the canonical translation initiation factors) as well as unspliced transcripts. The granules consist of 5´ends of polycistronic transcripts with their respective RNA binding proteins, that stick out of the pores to the cytoplasmic site. One major focus of the lab is the further characterisation of this highly unusual nuclear export pathway in trypanosomes. In particular, we want to understand the mechanisms of nuclear export control.
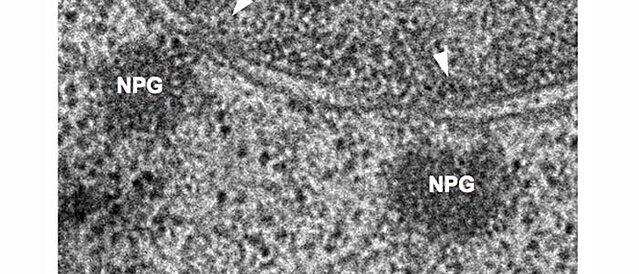 Nuclear periphery granules (NPGs), visualised by electron microscopy.
Nuclear periphery granules (NPGs), visualised by electron microscopy.
Publications
Goos C, Dejung M, Wehman AM, Meyer-Natus E, Schmidt J, SunterJ , Engstler M, Butter F and Kramer S. (2019) Trypanosomes can initiate nuclear export co-transcriptionally. Nucleic Acid research, in press PMID:30418648
Zoltner, M., Krienitz, N., Field, M. C. & Kramer, S (2018). Comparative proteomics of the two T. brucei PABPs suggests that PABP2 controls bulk mRNA. PLoS Necl Trop Dis 12, e0006679. PMID:30040867
Kramer S (2017) Simultaneous detection of mRNA transcription and decay intermediates by dual colour single mRNA FISH on subcellular resolution. Nucleic Acids Res 45: e49 PMID:27940558
Goos C, Dejung M, Janzen CJ, Butter F and Kramer S (2017) The nuclear proteome of Trypanosoma brucei. PLoS One 12(7):e0181884. PMID:28727848
Kramer S, Piper S, Estevez A, Carrington M. (2016). Polycistronic trypanosome mRNAs are a target for the exosome. Mol Biochem Parasitol 205:1–5. PMID:26946399
Kramer S, Bannerman-Chukualim B, Ellis L, Boulden EA, Kelly S, Field MC, Carrington M. (2013). Differential localization of the two T. brucei poly(A) binding proteins to the nucleus and RNP granules suggests binding to distinct mRNA pools. PLoS One 8: e54004. PMID:23382864
Kramer S, Marnef A, Standart N, Carrington M. (2012). Inhibition of mRNA maturation in trypanosomes causes the formation of novel foci at the nuclear periphery containing cytoplasmic regulators of mRNA fate. J Cell Sci 125: 2896-2909. PMID:22366449