3DART-VeSElecT
3D Automated Reconstruction Tool for Vesicle Structures of Electron Tomograms.
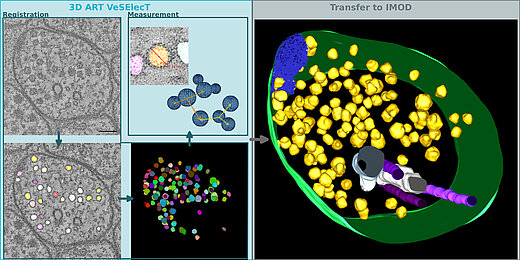
Automatic image reconstruction is critical to cope with steadily increasing data from advanced microscopy. We describe here the Fiji macro 3D ART VeSElecT which we developed to study synaptic vesicles in electron tomograms. We apply this tool to quantify vesicle properties (i) in embryonic Danio rerio 4 and 8 days past fertilization (dpf) and (ii) to compare Caenorhabditis elegans N2 neuromuscular junctions (NMJ) wild-type and its septin mutant (unc-59(e261)). We demonstrate development-specific and mutant-specific changes in synaptic vesicle pools in both models. We confirm the functionality of our macro by applying our 3D ART VeSElecT on zebrafish NMJ showing smaller vesicles in 8 dpf embryos then 4 dpf, which was validated by manual reconstruction of the vesicle pool. Furthermore, we analyze the impact of C. elegans septin mutant unc-59(e261) on vesicle pool formation and vesicle size. Automated vesicle registration and characterization was implemented in Fiji as two macros (registration and measurement). This flexible arrangement allows in particular reducing false positives by an optional manual revision step. Preprocessing and contrast enhancement work on image-stacks of 1nm/pixel in x and y direction. Semi-automated cell selection was integrated. 3D ART VeSElecT removes interfering components, detects vesicles by 3D segmentation and calculates vesicle volume and diameter (spherical approximation, inner/outer diameter). Results are collected in color using the RoiManager plugin including the possibility of manual removal of non-matching confounder vesicles. Detailed evaluation considered performance (detected vesicles) and specificity (true vesicles) as well as precision and recall. We furthermore show gain in segmentation and morphological filtering compared to learning based methods and a large time gain compared to manual segmentation. 3D ART VeSElecT shows small error rates and its speed gain can be up to 68 times faster in comparison to manual annotation. Both automatic and semi-automatic modes are explained including a tutorial.
Tutorial
Together with the macros "3DART_VeSElecT_RegistVesicle" and "3DART_VeSElecTMeasureVesicle" we provide a Fiji version (1.51g for Linux operating systems) on this homepage.
We succsessfully tested the macros for Fiji version 1.51g for different operating systems (Linux, Windows and Macintosh), which is the latest version so far.
Still, it is recommended to use the here provided version in combination with 3D ART VeSElecT for two reasons. On the one hand, this version already includes an additional plugin (3D ImageJ Suite), on the other hand, we can not guarantee the flawless function of the macro for other versions, e.g. in case of updates.
If you would like to test 3D ART VeSElecT we provide a test stack and a user description for download.
Downloads
Macros:
Click HERE to download the macros "3DART_VeSElecT_RegistVesicle" and "3DART_VeSElecTMeasureVesicle" as zip file.
User description:
Click HERE to download the user description "How_to_use_3D_ART_VeSElecT".
Test stack:
Click HERE to download the test_stack.tif of an electron microscopic tomogram.
Fiji:
Click HERE to download FIJI (1.51g for Linux operating systems, includes 3D ImageJ Suite).
Notice: If a different FIJI version than the one which is provided above is used, additionally the "3D ImageJ Suite" plugin has to be installed (for more information about 3D ImageJ Suite click HERE).
For more information about FIJI visit : www.fiji.sc
Classification Macro
The classification macro for FIJI/ImageJ is designed for classification of dense core and clear core vesicles in electron tomograms.
Abstract
Synaptic vesicles (SVs) are a key component of neuronal signaling and fulfil different roles depending on their composition. In electron micrograms of neurites, two types of vesicles can be distinguished by morphological criteria, the classical “clear core” vesicles (CCV) and the typically larger “dense core” vesicles (DCV), with differences in electron density due to their diverse cargos. Compared to CCVs, the precise function of DCVs is less defined. DCVs are known to store neuropeptides, which function as neuronal messengers and modulators. In C. elegans, they play a role in locomotion, dauer formation, egg-laying, and mechano- and chemosensation. Another type of DCVs, also referred to as granulated vesicles, are known to transport Bassoon, Piccolo and further constituents of the presynaptic density in the center of the active zone (AZ), and therefore are important for synaptogenesis.
To better understand the role of different types of SVs, we present here a new automated approach to classify vesicles and to quantify their properties from electron tomograms. We combine machine learning with an extension of our previously developed vesicle segmentation workflow, the ImageJ macro 3D ART VeSElecT, to reliably distinguish CCVs and DCVs using image-based features. We apply this method to electron tomograms of the C. elegans NMJs, and find an increased fraction of DCVs as well as a higher mean distance between DCVs and AZs in dauer larvae when compared to adult hermaphrodites. Our machine learning approach could be applied to study properties of different synaptic vesicle pools in electron tomograms of diverse model organisms.
Downloads
Software
3D ART VeSElecT 2 (updated version of 3D ART VeSElecT)
classification macro
python script
Tutorials
For application instructions for the classification macro download:
How_to_use_the_classification_macro
If you want to retrain the support vector machine algorithm, download the following instructions:
How_to_use_the_python_script
Test data
StackSegmented
StackDuplicateScale
Update ImageJ classification macro
This update reads-in scale information from StackDuplicateScale and puts this on new created figures so that the z-distance is accurately given. Moreover SVM parameters have been updated.
DCV_CCV_separation.ijm
References
Kaltdorf KV, Theiss M, Markert SM, Zhen M, Dandekar T, Stigloher C, et al. (2018) Automated classification of synaptic vesicles in electron tomograms of C. elegans using machine learning. PLoS ONE 13(10): e0205348.
(Download paper as PDF, click HERE)
Please cite this reference if you use 3D ART VeSElecT:
Kaltdorf, K.V. et. al., 2017. FIJI Macro 3D ART VeSElecT: 3D Automated Reconstruction Tool for Vesicle Structures of Electron Tomograms., pp. 1-21.
(Download Paper as PDF, click HERE)
FIJI is provided by:
Schindelin, J. et al., 2012. Fiji: an open-source platform for biological-image analysis. Nature methods, 9(7), pp.676–82. Available at: http://dx.doi.org/10.1038/nmeth.2019
3D ImageJ Suite:
J. Ollion, J. Cochennec, F. Loll, C. Escudé, T. Boudier. (2013) TANGO: A Generic Tool for High-throughput 3D Image Analysis for Studying Nuclear Organization. Bioinformatics 2013 Jul 15;29(14):1840-1.