TRR221
Basis für unser Datenmanagement sind die Forschungsdatenmanagement Grundsätze unserer Universität
Forschungsdaten sind alle Daten und Proben, die im Verlauf eines Forschungsprozesses z. B. durch Quellenforschungen, Experimente, Messungen, Erhebungen oder Befragungen entstehen, entwickelt oder ausgewertet werden.
Forschungsdatenmanagement bezeichnet alle Prozesse innerhalb des Forschungsdatenzyklus. Es beinhaltet die Erzeugung, Dokumentation, Analyse und Aufbereitung, Nutzung, Archivierung, Veröffentlichung, Nachnutzung ggf. bis hin zur Löschung von Forschungsdaten.
Hilfreich ist die Erstellung eines Datenmanagementplans bereits bei der Projektplanung. Einige Forschungsförderer schreiben dies vor und erwarten abschließend eine Open-Access-Veröffentlichung der Forschungsdaten. Ausführliche Informationen dazu erhalten Sie in unseren Handlungsempfehlungen.
Geeignete Forschungsdatenrepositorien sowohl für die Suche nach Forschungsdaten als auch für die Veröffentlichung von Forschungsdaten sind in der Registry of Research Data Repositories (re3data.org) gelistet.
Für die Wissenschaftler der Universität Würzburg bündeln das Servicezentrum Forschung und Technologietransfer (SFT), das Research Advancement Centre (RAC), die Universitätsbibliothek (UB) und das Rechenzentrums (RZ) ihre Kompetenzen.
Quelle: https://www.uni-wuerzburg.de/rdm/
The significant changes in the landscape of higher education, research, and knowledge and technology transfer that digital transformation has brought about have caused a growing need for effective research data management (RDM). Julius-Maximilians-Universität Würzburg (JMU) provides guidance and support for researchers that are involved in the acquisition of third-party funding, the implementation of research projects, and other aspects of RDM. The University encourages its researchers to involve all collaborators that contribute to a research project in RDM from an early stage and to develop a data management plan (DMP). At the planning stage of any research project, you should devote particular attention to developing a strategy for the management, publication, and use of the data generated by that project. To ensure the success of your project and the re-usability of the generated data, you should also make arrangements for the long-term storage of that data. Having proper data management procedures in place ensures long-term protection from data loss that is independent of the individuals involved. To facilitate access to and use of your research data, you should use transparent archiving structures and provide contextual information for that data. Please also bear in mind that pure data publications are considered fully-fledged publications that can help you make your work more visible and raise your research profile: Your data will be assigned a DOI; this means that they will be citable without having been discussed in journal articles and can be used in some forms of citation analysis, lists of publications, etc.
Quelle: https://www.uni-wuerzburg.de/en/data/good-practice-in-research-data-management/
Forschungsdatenmanagement (FDM) nimmt aufgrund der rasant voranschreitenden Digitalisierung in Forschung, Lehre sowie im Wissens- und Technologietransfer eine immer zentralere Rolle im Forschungsgeschehen ein. An der JMU existieren bereits Unterstützungsstrukturen für Akquise und Durchführung von Forschungsprojekten und weitere Aspekte des FDM. Grundsätzlich wird dazu geraten, alle Kooperationspartner, die zu einem Forschungsprojekt beitragen, möglichst frühzeitig in das FDM einzubinden und einen Datenmanagementplan (DMP) aufzustellen. Das Vorgehen im Umgang mit gewonnenen Daten und mit deren Veröffentlichung und Verwertung sollte integraler Bestandteil der Planung eines Forschungsprojektes sein. Die langfristige Speicherung von Daten beinhaltet ebenfalls eine Reihe von Gesichtspunkten und Entscheidungen, die für den erfolgreichen Projektverlauf und die Wiederverwertung der Forschungsdaten wichtig sind. Ein sinnvolles Datenmanagement sichert Ihre Daten langfristig und personenunabhängig vor Verlust. Aufgrund nachvollziehbarer Archivierungsstrukturen und gespeicherter Kontextinformationen bleiben die Daten auch auf lange Sicht leichter auffindbar und nutzbar. Schließlich sind die positiven Auswirkungen reiner Datenpublikationen nicht zu unterschätzen. Sie gelten als eigenständige Publikationen, so dass Wissenschaftler und Wissenschaftlerinnen ihre Forschungsleistungen sichtbarer machen und ihre Reputation steigern können: Die veröffentlichten Daten werden durch Zuweisung einer DOI bereits ohne eine Besprechung in wissenschaftlichen Zeitungsartikeln zitierfähig und können z. B. in manchen Formen von Zitationsanalysen oder in Publikationslisten verwendet werden.
Quelle: https://www.uni-wuerzburg.de/rdm/
current results:
Functional genomics identifies AMPD2 as a new prognostic marker for undifferentiated pleomorphic sarcoma
(funder: Land of Bavaria)
Vaccinia virus injected human tumors: oncolytic virus efficiency predicted by antigen profiling analysis fitted boolean models
(funder: Land of Bavaria, TRR221 acknowledged)
Transregio 221 "Transplant-gegen-Wirt und Transplant-gegen-Leukämie Immunreaktionen"
Homepage Transregio 221 in Regensburg
Im Sonderforschungsbereich Transregio 221 wollen Immunmediziner und Wissenschaftler der Universitäten Regensburg, Erlangen und Würzburg und der dortigen Universitätsklinika gemeinsam Wege finden, die allogene Stammzelltransplantation zukünftig sicherer und erfolgreicher zu machen, indem Krankheitsrückfälle vermieden und Transplantationskomplikationen vermindert werden.
GvL
Für Patienten mit einer Leukämie- oder Lymphomerkrankung ist die Chemotherapie die wichtigste Behandlungsform. Allerdings bildet sich bei einem Teil der Patienten die Erkrankung trotz Chemotherapie nicht zurück oder sie tritt nach einem ersten Therapieerfolg erneut auf. In vielen Fällen ist dann die allogene Blutstammzelltransplantation die einzige Heilungsmöglichkeit. Deren Wirksamkeit beruht darauf, dass gesunde Stammzellen und Immunzellen eines Spenders die Blutbildung des Patienten ersetzen sowie noch vorhandene Leukämiezellen bzw. Lymphomzellen zerstören. Diese wichtige Leistung des Spenderimmunsystems wird als Transplantat-gegen-Leukämie-Effekt (graft-versus-leukemia, GvL) bezeichnet. Der GvL-Effekt ist allerdings nicht bei allen Patienten ausreichend stark, um einen Rückfall der Leukämie- oder Lymphomerkrankung zu verhindern.
Data Management Plan (DMP)
Collaborative Research Centre (Transregio) 221: Graft versus host disease / Graft versus leukemia “GvHD / GvL”
1. Data repository - our fundamental and basic task
Data Management and Data Integration starts with basic requirements (interface, databank model, access policy), develops software solutions for different data types and user wishes and involves various and large data sets and their curation (lot of work). To achieve a new level of integration is exciting but has the challenge to connect various different entities together. For our specific question, GvHD /GvL, we envision a systems biological repository, first established by sorting the INF data (10s of Terabytes processed data; in particular all publication relevant data; raw data stay with the PIs of the CRC) according to different GvHD /GvL models, but profiting from the database structure to have sorting both regarding the different GvHD /GvL models and types of data. Fine-structuring of the data will be done by immunological and cell biology criteria and hence exploring the tissue and specific experimental conditions. This will rely on gene onthology and similar molecular classification categories according to available experimental meta-data and raw data. The added value from comparing and integrating the different INF data-sets and types (e.g. imaging plus high resolution RNAseq) includes cell-specific resolution of the GvHD /GvL processes and allows investigation of different immune responses.
Investigation of GvHD /GvL with state-of-the art technologies creates a lot of data, in particular regarding image data and large scale transcriptomics and proteomics data sets.
For this Big Data wave the SFB, relying on its INF project, will provide suitable data management starting from a data management plan. We will focus on long term maintenance of all publication-relevant data as well as proper deposition of primary data and required raw data. We have long-term experience in providing such frameworks in different INF projects of collaborative research centers as well as providing bioinformatic platforms for maintenance and analysis of Big Data from Omics technologies and imaging. We will store all published final data of the GvHD /GvL CRC including image data, but excluding raw data as well as patient data (for all clinical data, in particular patient data, the different university clinics biometricians are responsible and should be contacted). For efficient data storage, not just the data but also the experimental meta-data are important.
Vision: Our vision is data integration for better understanding GvHD/GvL, visualize and explore synergistically the different OMICs data sets against each other including also large-scale sequence data including single sequencing data. Considering the different compartments of the GvHD /GvL processes, we will strive to provide a first basis to connect all stored molecular entities, cells or tissue events into an integrated picture. Moreover, imaging data modalities include advanced fluorescence microscopy such as confocal, time-lapse, intravital, or light-sheet microscopy as well as corresponding image processing software. An additional level of complexity are functional assays on immune cells, blood and biochemical data.
On this more sophisticated analysis of the GvHD /GvL process may be done (data warehousing; data type and onthology classification; molecular analysis and classification; systems biological modelling), in particular a thorough classification of involved immune cells and tissue, noting host-specific immune cells and tissues with different immune make-up as well as graft-specific immune cells and responses and further entities, in particularly residual disease and leukemic cells. Also here sub-classification according to OMICs and further data is critical as well as modelling of involved interactions, and higher level systems biological processes, the higher levels are all cellular processes in graft, host and tumor cells contributing to the interactions in GvHD /GvL.
It is our vision that our data-collection, integration and joined analysis with all other groups in the context of the CRC ultimately provides an important blue print of central disease-relevant processes in GvHD /GvL.
Of course, this is really time intensive and requires direct, personal collaboration and dedicated students at least at the master level. However, for careful selected topics and questions this is of course possible even with our limited resources. we are very open to this and hope with our vision to catalyze the collaborative spirit in the CRC 221. Please contact us!
2. Our help in Data storage and exchange
We will help and advice in storing your primary data at your site (including: advise on storage media, estimate of storage needs, and help in obtaining direct financial support from the CRC for storage requirements)
[We have here a central data exchange service and links for the CRC 221: …]
3. Support in Data processing and Data analysis
3.1. Direct access and help in data analysis
In our INF project we provide lots of different analysis software for all sorts of data analysis, starting from sequence analysis to protein domain and structure analysis, analysis of RNA including regulatory motifs, RNA secondary structure and RNA-based phylogenetic analysis.
https://www.biozentrum.uni-wuerzburg.de/bioinfo/computing/
3.2. Preprocessing and large-scale data analysis pipelines
Moreover, for processing of data, we provide whole analysis flows
Fill in: NGS data analysis flow, simple annotation analysis flow;
Network analysis flow: Metabolic network analysis
Network analysis flow: Signalling network analysis
(add our Tim Breitenbach paper where you have TR221 acknowledged)
RNA analysis flow (Max Fuchs pipeline and paper)
Gene expression analysis flow, cancer specific analysis questions (Meik Kunz paper)
3.3. Direct tuition, lectures and courses
3.3.1. Books:
First of all, we have a whole book on bioinformatics explaining you the fundamentals for your analysis:
Springer Link (our German bioinformatics book)
Moreover, there are two very good English textbooks:
Introduction to bioinformatics (Prof. Arthur Lesk)
Systems biology (Prof. Edda Klipp & coworkers)
3.3.2. Courses & Meetings:
Retreat TR221 7./8.12.2018 Schwarzenfeld
Our summer school infection biology
The summer schools at the CCTB
3.3.3. Publications on bioinformatics
General publications by the Dandekar groups
Our TR221-specific publications:
Vaccinia virus injected human tumors: oncolytic virus efficiency predicted by antigen profiling analysis fitted boolean models (2019). Cecil A, Gentschev I, Adelfinger M, Dandekar T, Szalay AA. Bioengineered. 2019 Dec;10(1):190-196. doi: 10.1080/21655979.2019.1622220.
Functional genomics identifies AMPD2 as a new prognostic marker for undifferentiated pleomorphic sarcoma (2018). Orth MF, Gerke JS, Knösel T, Altendorf-Hofmann A, Musa J, Alba-Rubio R, Stein S, Hölting TLB, Cidre-Aranaz F, Romero-Pérez L, Dallmayer M, Baldauf MC, Marchetto A, Sannino G, Knott MML, Wehweck F, Ohmura S, Li J, Hakozaki M, Kirchner T, Dandekar T, Butt E, Grünewald TGP. Int J Cancer. 2019 Feb 15;144(4):859-867. doi: 10.1002/ijc.31903. Epub 2018 Dec 4.
Data management implementation
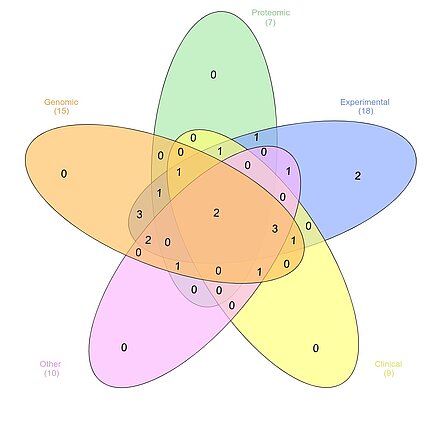
Data management plan for different working protocols, the group numbers are shown in the figure, the most demanding data management is Genomic/Transcriptomic, including group A1, A3, A6, B1, B3, B4, B6, B7, B8, B9, B10, B12, B13, Z2, INF. Second is experiment document and protocol, including group A1, A2, A3, A4, A5, A6, B1, B3, B4, B6, B7, B8, B9, B10, B11, B12, Z1, Z2. Next is Clinical data A3, A6, B3, B6, B9, B10, B12, B13, Z1. Proteomic data management (A1, A3, A4, B9, B12, Z1, INF) and others (A3, A6, B6, B7, B9, B11, B10, B13, Z2, INF) including long-term data warehousing and pathway analysis are the last.
The current data server is located at the computer center in the University of Würzburg. The starting quota for the data cloud is 2 TB (used). The workbench platform is currently implemented in the same server for a rapid data management. In the future, they will be separated for a safer management.