TRR240
Basis für unser Datenmanagement sind die Forschungsdatenmanagement Grundsätze unserer Universität
Forschungsdaten sind alle Daten und Proben, die im Verlauf eines Forschungsprozesses z. B. durch Quellenforschungen, Experimente, Messungen, Erhebungen oder Befragungen entstehen, entwickelt oder ausgewertet werden.
Forschungsdatenmanagement bezeichnet alle Prozesse innerhalb des Forschungsdatenzyklus. Es beinhaltet die Erzeugung, Dokumentation, Analyse und Aufbereitung, Nutzung, Archivierung, Veröffentlichung, Nachnutzung ggf. bis hin zur Löschung von Forschungsdaten.
Hilfreich ist die Erstellung eines Datenmanagementplans bereits bei der Projektplanung. Einige Forschungsförderer schreiben dies vor und erwarten abschließend eine Open-Access-Veröffentlichung der Forschungsdaten. Ausführliche Informationen dazu erhalten Sie in unseren Handlungsempfehlungen.
Geeignete Forschungsdatenrepositorien sowohl für die Suche nach Forschungsdaten als auch für die Veröffentlichung von Forschungsdaten sind in der Registry of Research Data Repositories (re3data.org) gelistet.
Für die Wissenschaftler der Universität Würzburg bündeln das Servicezentrum Forschung und Technologietransfer (SFT), das Research Advancement Centre (RAC), die Universitätsbibliothek (UB) und das Rechenzentrums (RZ) ihre Kompetenzen.
Quelle: https://www.uni-wuerzburg.de/rdm/
The significant changes in the landscape of higher education, research, and knowledge and technology transfer that digital transformation has brought about have caused a growing need for effective research data management (RDM). Julius-Maximilians-Universität Würzburg (JMU) provides guidance and support for researchers that are involved in the acquisition of third-party funding, the implementation of research projects, and other aspects of RDM. The University encourages its researchers to involve all collaborators that contribute to a research project in RDM from an early stage and to develop a data management plan (DMP). At the planning stage of any research project, you should devote particular attention to developing a strategy for the management, publication, and use of the data generated by that project. To ensure the success of your project and the re-usability of the generated data, you should also make arrangements for the long-term storage of that data. Having proper data management procedures in place ensures long-term protection from data loss that is independent of the individuals involved. To facilitate access to and use of your research data, you should use transparent archiving structures and provide contextual information for that data. Please also bear in mind that pure data publications are considered fully-fledged publications that can help you make your work more visible and raise your research profile: Your data will be assigned a DOI; this means that they will be citable without having been discussed in journal articles and can be used in some forms of citation analysis, lists of publications, etc.
Quelle: https://www.uni-wuerzburg.de/en/data/good-practice-in-research-data-management/
Forschungsdatenmanagement (FDM) nimmt aufgrund der rasant voranschreitenden Digitalisierung in Forschung, Lehre sowie im Wissens- und Technologietransfer eine immer zentralere Rolle im Forschungsgeschehen ein. An der JMU existieren bereits Unterstützungsstrukturen für Akquise und Durchführung von Forschungsprojekten und weitere Aspekte des FDM. Grundsätzlich wird dazu geraten, alle Kooperationspartner, die zu einem Forschungsprojekt beitragen, möglichst frühzeitig in das FDM einzubinden und einen Datenmanagementplan (DMP) aufzustellen. Das Vorgehen im Umgang mit gewonnenen Daten und mit deren Veröffentlichung und Verwertung sollte integraler Bestandteil der Planung eines Forschungsprojektes sein. Die langfristige Speicherung von Daten beinhaltet ebenfalls eine Reihe von Gesichtspunkten und Entscheidungen, die für den erfolgreichen Projektverlauf und die Wiederverwertung der Forschungsdaten wichtig sind. Ein sinnvolles Datenmanagement sichert Ihre Daten langfristig und personenunabhängig vor Verlust. Aufgrund nachvollziehbarer Archivierungsstrukturen und gespeicherter Kontextinformationen bleiben die Daten auch auf lange Sicht leichter auffindbar und nutzbar. Schließlich sind die positiven Auswirkungen reiner Datenpublikationen nicht zu unterschätzen. Sie gelten als eigenständige Publikationen, so dass Wissenschaftler und Wissenschaftlerinnen ihre Forschungsleistungen sichtbarer machen und ihre Reputation steigern können: Die veröffentlichten Daten werden durch Zuweisung einer DOI bereits ohne eine Besprechung in wissenschaftlichen Zeitungsartikeln zitierfähig und können z. B. in manchen Formen von Zitationsanalysen oder in Publikationslisten verwendet werden.
Quelle: https://www.uni-wuerzburg.de/rdm/
German Overview
Our Support Project (INF)
Besides infrastructure for data storage, pointers and exchange for the whole CRC we analyze, process and integrate the public and protected CRC-TRR240 data in our easy to navigate Virtual Platelet Platform ViPP. This includes scientific simulations as well video-tutorials and other teaching material (Virtual Platelet Classroom “ViP-Class”). It builds on our previous work on molecular data integration with a platelet web systems biological workbench, including metabolic, regulatory and dynamical models and simulations for platelet activation and inhibition. This is complemented by imaging data and advanced image analysis tools: Data of Electron Tomograms, and different fluorescence approaches such as confocal, time-lapse, intravital, or light-sheet microscopy. ViPP integrates molecular, pathway/compartment and intercellular/tissue information in a novel way. Corresponding molecular, network and image processing software, protocols and pipelines will be part of ViPP. We will follow the vision of an integrated, new and powerful research environment, where omics data provide highest resolution, blending in with imaging data on various scales including data on cellular processes and responses as well as simulations. The first period will explore this new level of data integration. The long term goal is active participation and ultimately a fine grained Virtual Research Environment for platelets.
Current state of understanding To date
there are two major challenges regarding Big Data in both genome genome-scale technologies (“-omics”) and imaging: First, the massive amount of data need to be stored, retrieved, and ‘directly’ accessible, second the data need to be analysed, and resulting meta data or models need to be made available to the scientist involved. One start requirement particular for reliable –omics data is Standard Operation Procedures (SOPs) to provide a framework for data comparability and reproducibility.
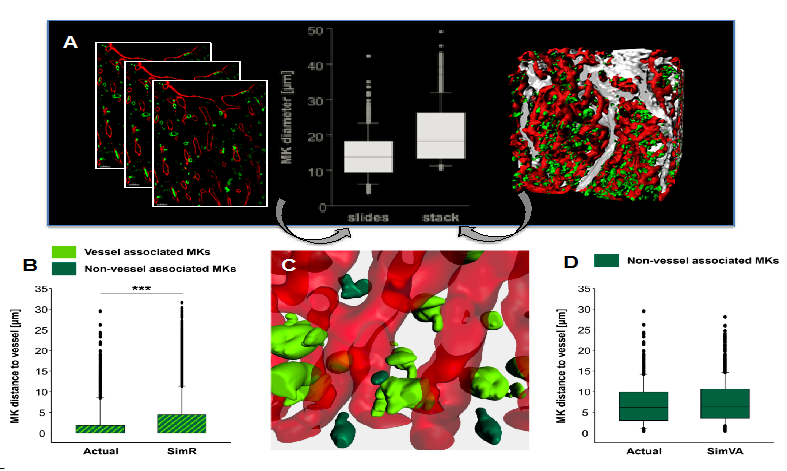
Figure: Mouse sternum bone marrow images ((A, KH, unpublished) and simulations (B-D, derived from VI): (A) shows densely packed vasclature (red), MKs (green) and bone (grey) in 2D (left) and 3D (right) with analysis of examplied slides and corresponding stack( middle). (B)-(D) shows simulated MK distribution (n=6 simulations) using blood vessels (red) and MKs (green) from segmented imaging data. (B) The average distance of random distributed MKs to the vessel (SimR) is increased compared to actual data. ***, p<0.001. (C) Simulated vessel-associated MKs (light green) versus non-vessel-associated MKs (dark green). (D) MKs display a vessel-biased random distribution: Random simulation of only the non-vessel associated MK population results in MK-vessel distances matching the 3D imaging data. This is complemented by analysis of the involved molecular components and networks (TD), here focusing on MK and collagen IV or I interaction in the extracellular matrix, critical for platelet generation by MKs.
Principal investigator(s)
Thomas Dandekar, Prof. Dr. med. Lehrstuhl für Bioinformatik, Biozentrum Universität Würzburg Am Hubland 97074 Würzburg Phone: +49 931 31-84551 E-mail: dandekar@biozentrum.uni-wuerzburg.de | Katrin G. Heinze, Prof. Dr. rer. nat. Bio-Imaging-Zentrum und Rudolf-Virchow-Zentrum Universität Würzburg Josef-Schneider-Str. 2 / D15 97080 Würzburg Phone: +49 931 31-84214 E-mail: katrin.heinze@virchow.uni-wuerzburg.de |