JimenaC
Our latest software from the Jimena suite (Karl and Dandekar, 2015; link) is JimenaC (C for "compare cooperative system changes").
JimenaC is generally applicable to study dynamic change and cooperative pathway activation as well as switching behavior in protein-protein networks. JimenaC and its code are optimized to model the activity changes of pathways involved in such a network, interpolating with different functions between the on and off states of the protein interaction network according to its Boolean network logic. In particular, JimenaC allows to model system change, cooperative pathway change and system control in interaction networks. Validation data are easily incorporated and can easily be compared to simulation results.
Thus, both in bacterial (B.subtilis, S.aureus) and in fungal systems (C. albicans, A. fumigatus) it proofed useful to model dynamic network responses. We study with it biofilm formation, quorum sensing in prokaryotes and infection biology, fungal growth as well as immune cell responses in fungal interactions. Finally, an important further application area extend the human cellular models to simulate and study cancer cells as well as synthetic organisms such as the oncolytic virus and the resulting system effects and cooperative pathway effects)
JimenaC is a Java genetic regulatory network simulation framework which focuses on computational efficacy and a modularized architecture to facilitate the development and testing of new algorithms and models surrounding GRNs. It features
- Network import from yED graph files (Example 1: Download (source), Example 2: Download)
- Full support of the exponential interpolation model implemented in SQUAD
- Full support of the polynomial interpolation models implemented in Odefy, including a significantly better implementation of the interpolation algorithm
- Several discrete update schemes
- Perturbation support in all models
- Several algorithms for the search of steady stable states in all models (fully multithreaded)
- In development: Network stability analyses, etc.
It is developed by the Department of Bioinformatics of the University of Würzburg and supervised by Thomas Dandekar (head of the department).
Availability
JimenaC is publically avaible via here
It requires an environment of JRE version higher than 11 to run it,
in windows, simply click it after you have installed JRE. In linux, you need to "java -jar jimenaC.jar" to run it.
Network can be edited with yed software (URL:https://www.yworks.com/products/yed/download).
DEMO files
demo model (interaction_bacillus.graphml) is available here.
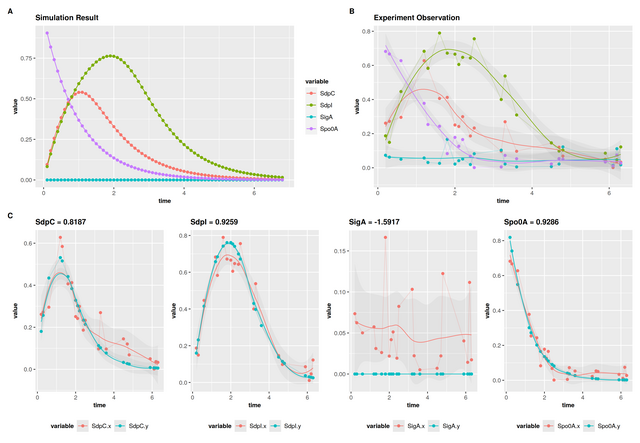
simulation result in a tsv (tab-separated values) format is here. The file can be generated directly using JimenaC export function, however, please edit it using excel or spreadsheet to remove all the protein columns you are interested to minimize the number of proteins for the downstream validation and comparison. e.g., in the demo file, you can find only four interesting proteins. We suggest you choose four to sixteen proteins.
experiment observation for validation in a tsv format is here.
the validation R-script is available here. Please change the two file names according to your input file. Note the script requires four R library, dplyr, reshape2, ggplot2 and patchwork, you might need to install it before you run the script.
Application
Cooperative changes are critical to achieve switches in systems. We offer molecular and modelling tools to design these including four key principles to achieve cooperativity in different systems: Efficient task distribution, cooperative planning and perception together with information exchange between components. We calculate and engineer cooperative changes in proteins using light as switching signal. We monitor and predict cooperative pathway changes in B. subtilis biofilm differentiation and oncolytic vaccinia virus acting on cancer cells applying Boolean semi-quantitative simulations. Future extensions include smart nanobiotechnology, active DNA storage and improved oncolysis in cancer. Technical and cellular process switches are designed and compared using GoSynthetic database. We demonstrate how technical systems cooperate using multi-level communication and self-organization in networked robots and satellites.
Update
- Fixed a bug where duplicate stable states were not recognized
- Multiple result windows can now be open at the same time
- States (e.g. stable states) can now be copied (Ctrl+C) from any window (e.g. stable state search) and entered into the nodes window (Ctrl+V)
- The charts window now exports the time sequence data as a tab-separated table (compatible with common spreadsheet software)
- Network states can now be loaded from text files and saved to them
- The user may now choose whether to include the discrete stable states in a random search for stable states
- Stable states can now be exported to a file and to the clipboard
Network input format
Activating influences are modeled using the yED standard arrow tip, inhibiting influences using any other tip (e.g. a diamond). Boolean nodes are modeled using nodes with the captions AND, OR, NOT, &&, ||, !. Cf. the examples above.
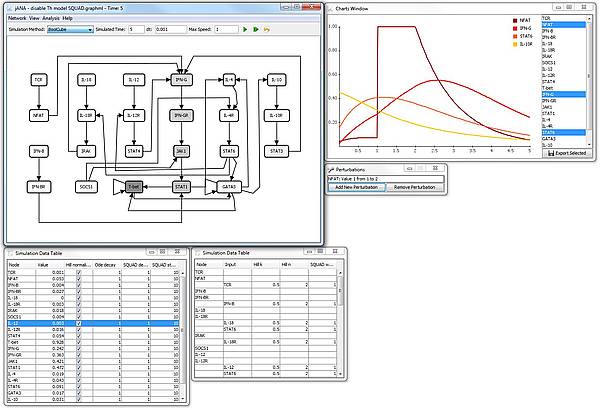
Screenshot
The screenshow shows a simulation of the T-helper network in the jIMENA GUI (which is not yet included in the release). The simulation was started from random values and the NFAT node was set to 1 between the time indices 1 and 2 by a perturbation.
Compare Jimena Simulation to experimental data
Exampole data: See this exel file. Just load up your experimental data file and then JimenaC will compare the experimental data to the simulation.
Contact
Prof. Thomas Dandekar
Dandekar group, Functional Genomics and Systems Biology department,
Lehrstuhl für Bioinformatik
Biocerter, University of Wuerzburg Am Hubland,
D-97074 Würzburg
Tel.: +49 (0)931 31 84551
Fax: +49 (0)931 31 84552
Ackowledgements
- JavaBDD (GNU LGPL) by John Whaley (http://javabdd.sourceforge.net) is used as a BDD framework.
- Icons (GNU LGPL) by Oxygen Team (http://www.iconarchive.com/artist/oxygen-icons.org.html)
- Several known GRN models have been implemented in Jimena. Cf. the papers for a comprehensive list.
- Support by DFG (Da 208/12-1) is gratefully acknowledged.