Isotop software
Isotopo
Isotopo is an application with the ability of performing quantitative mass spectrometry to readily mixtures of materials labeled with stable isotopes.
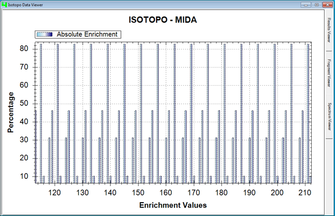
Isotopo is the ability of processing experimental isotopomers data (output of GC-MS) and provides obtained results (output) both in textual and visual formats, for better understanding and analysis. Processable (input) data consists of following elements i.e. Metabolite Information, Mass to Charge Ratio (M/Z) Values, Actual Relative Intensity (RI) Values and Standard Relative Intensity Values and Number of Carbon Atom Fragments. Isotopo is capable of analyzing three RIs value sets observed during GC-MS experiment against one M/Z value set with one Standard RI value set. The resultant (output) information is consists of following results i.e. Mass Calculations (Mo, M-1, M maximum), Natural Abundances (NA), Relative Abundances (RA), Fractional Molar Abundances (FRA), Absolute Enrichment, Mean RA, Standard Deviation RA and a Spectrum of calculated RA. It provides output of each processed RI along with average of three.
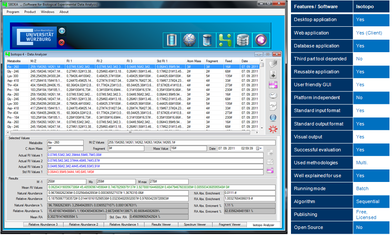

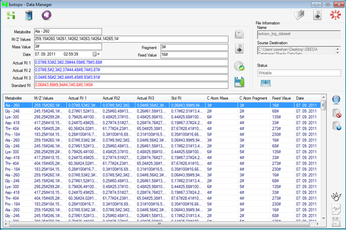
Isotopo also provides a file based and database management system for efficient experimental data manipulation and management.
Isotopo is implemented in Microsoft Visual Studio Dot Net (C#), following V-Model SDLC, integrating formal unified modelling language to scheme from different perspectives and incorporating human computer interaction guidelines, principles and patterns.
The freely available package for any non-commercial use includes a tutorial and allows robust data management of multiple labelling experiments
The most recent and available version of Isotopo provide various enhancements (as compared to LS-MIDA), including improvement in the matrix calculations, iterative refinement, quality control and mathematical validation, statistics on results, automatic data extraction / import / export and database management system.
Related Publications
- Ahmed Z., Saman Z., Huber C., Hensel M., Schomburg D., Münch R., Eylert E., Eisenreich W., Dandekar T. 'Isotopo' a Database Application for Facile Analysis and Management of Mass Isotopomer Data, Database: The Journal of Biological Databases and Curation, Oxford University Press, 2014.
- Dandekar T., Fieselmann A., Saman M. and Ahmed Z., Software Applications toward Quantitative Metabolic Flux Analysis and Visualization. Briefings in Bioinformatics, Oxford Journals, 15:1, 91-107, 2014.
- Ahmed Z, Saman Z, Dandekar T. Developing sustainable software solutions for bioinformatics using the “Butterfly” paradigm, F1000 Research, 3:71, 2014.
- Ahmed Z., Saman Z., Huber C., Hensel M., Schomburg D., Münch R., Eisenreich. W, and Dandekar T. Software LS-MIDA for Efficient Mass Isotopomer Distribution Analysis in Metabolic Modelling. BMC Bioinformatics, 14:218, 2013.
- Ahmed Z., Saman M., and Dandekar T. Computational Feature Performance and Domain Specific Architecture Evaluation of Software Applications towards Metabolic Flux Analysis. Recent Patents on Computer Science, 5:3, 165-176, 2012.
- Ahmed Z., Saman M., and Dandekar T., Formal UML Modelling of Isotopo, Bioinformatical Software for Mass Isotopomers Distribution Analysis. Software Engineering, SAP, 2:5, 147-159, 2012.
Conference Appearances:
- Ahmed Z., Eisenreich E., Dandekar T. Intelligent Metabolic Isotopomer Data Extraction, Classification, Standardization and Management, New England Database Summit (NEDB), CSAIL, the Massachusetts Institute of Technology (MIT), USA, 2013.
- Ahmed Z., Saman M., Eisenreich E., Dandekar T. Isotopo Towards Quantitative Mass Isotopomers Distribution Analysis using Spectral Data. 9th Symposium on the Practical Applications of Mass Spectrometry in the Biotechnology Industry (Mass Spec 2012), CASSS, Page 40, ID P-101, San Diego, CA USA, 2012.
- Ahmed Z., Saman M., Eisenreich E. Dandekar. Isotopo: Software towards Quantitative MIDA, Visualization and Data Management, 5th Annual New England Data Base Summit (NEDB), CSAIL, The Massachusetts Institute of Technology (MIT), USA, 2012.
Contacts
Dr. Zeeshan Ahmed
Department of Neurobiology and Genetics;
Biocenter, University of Wuerzburg.
Am Hubland 97074,
Wuerzburg Germany.
Prof. Dr. Thomas Dandekar
Department of Bioinformatics;
Biocenter, University of Wuerzburg.
Am Hubland 97074,
Wuerzburg Germany.