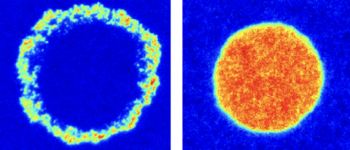
Bartossek T, Jones NG, Schäfer C, Cvitković M, Glogger M, Mott HR, Kuper J, Brennich M, Carrington M, Smith A-S, Fenz S, Kisker C, Engstler M. Structural basis for the shielding function of the dynamic trypanosome VSG coat, Nature Microbiology, doi:10.1038/s41564-017-0013-6.
Fenz S, Bihr T, Schmidt D, Merkel R, Sengupta K, Seifert U, Smith A.-S. (2017), Membrane fluctuations mediate lateral interaction between cadherin bonds, Nature Physics, doi: 10.1038/NPHYS4138.
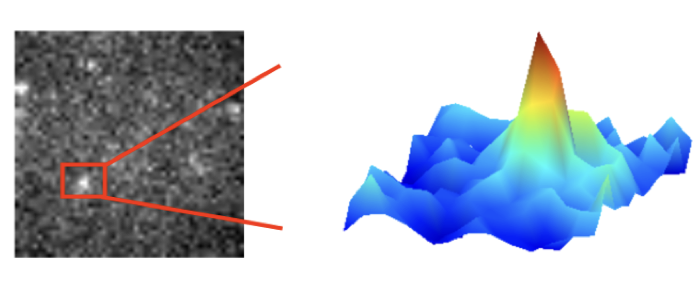
Fenz S, Smith A.-S., Monzel C. (2017) Measuring the invisible – Determining the size of growing nanodomains using the “inverse FCS”. Biophysical Journal, doi: 10.1016/j.bpj.2017.04.014.
Glogger M., Subota I., Pezzarossa A., Denecke A.-L., Carrington M., Fenz S.F., Engstler M. (2017) Facilitating trypanosome imaging, Experimental Parasitology, doi: 10.1016/ j.exppara.2017.03.010.
Glogger M, Stichler S, Subota I, Bertlein S, Spindler M-C, Tessmar J, Groll J, Engstler M, Fenz S (2017) Live-cell super-resolution imaging of intrinsically fast moving flagellates, Journal of Physics D: Applied Physics, doi: 10.1088/1361-6463/aa54eb
Beletkaia E, Fenz S, Pomp W, Snaar-Jagalska B. E., Hogendoorn P. W.C., Schmidt T (2016) CXCR4 signaling is controlled by immobilization at the plasma membrane, BBA: Molecular Cell Research, doi: 10.1016/j.bbamcr.2015.12.020
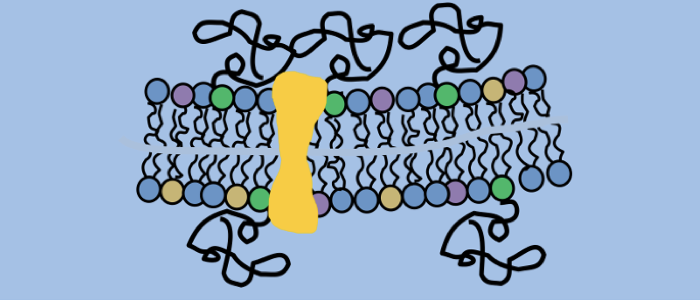
Hartel AJW, Glogger M, Jones NG, Abuillan W, Batram C, Hermann A, Fenz SF, Tanaka M, Engstler M (2016) N-glycosylation enables high lateral mobility of GPI-anchored proteins at a molecular crowding threshold. Nature Communications, doi: 10.1038/ncomms12870.