Novel virulence factors in Leishmania parasites
Leishmaniasis
Leishmania spp., a trypanosomatid parasite, is the causative agent of leishmaniasis, a neglected disease present worldwide in subtropical regions. There are three main manifestations of leishmaniases: visceral (the most serious form, which is almost always fatal without treatment), cutaneous (causing skin ulcers and scar tissue), and mucocutaneous (affecting mouth, nose and throat mucosa). With 700,000 to 1 million new cases annually worldwide, leishmaniasis primarily impacts impoverished individuals with compromised immunity due to poor living conditions. Limited access to medication due to financial constraints further contributes to disease prevalence, prompting efforts by the World Health Organization to promote medication access and support national leishmaniasis control programs. Climate change will most likely influence vector distribution, exacerbating the spread of the disese within many areas including Europe.
To be able to adapt to different host environments, Leishmania developed a complex life cycle. After transmission to vertebrates by phlebotomine sandflies, the parasite differentiate from extracellular promastigotes to intracellular amastigotes within host macrophages. This is an extremely hostile environment, but many sophisticated strategies enable the parasite to evade the host's defense systems. Many aspects of these processes are still elusive.
Leishmania host adaptation mechanisms
Leishmania parasites differentiate to an intracellular life cycle stage within the phagocytic cells of their vertebrate hosts. Numerous transcriptomic analyses have investigated host cell defense mechanisms and the parasite's responses to these processes. However, due to the prevalence of post-transcriptional regulation in kinetoplastids, transcriptomes often inadequately reflect the protein composition. To address this limitation, we conducted a systematic comparative in vitro study of the infection process using a quantitative proteomics approach.
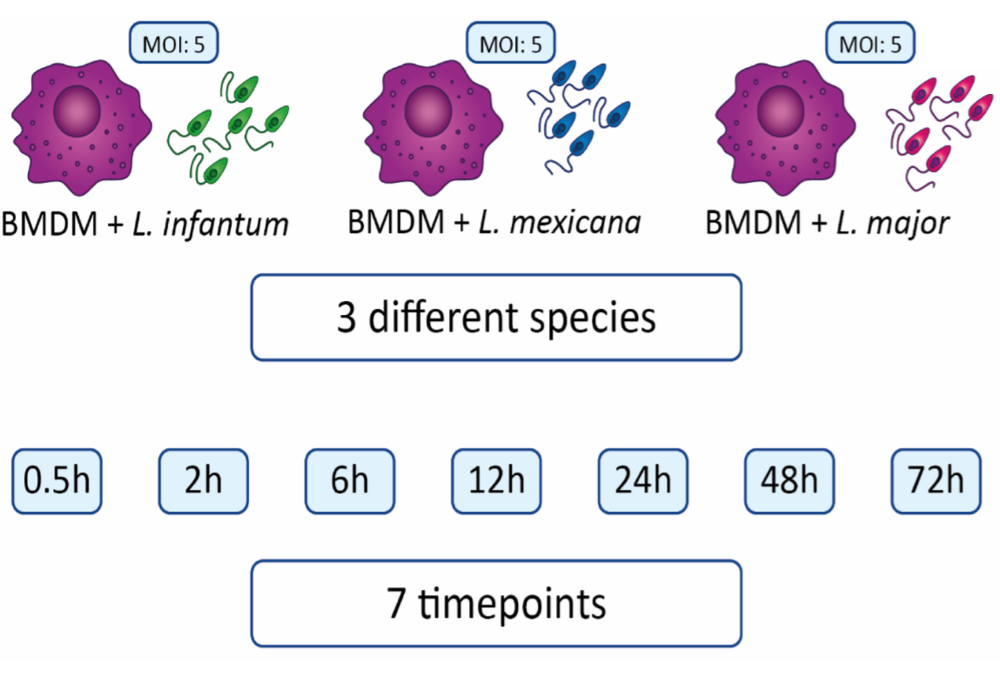
In our study, murine bone marrow-derived macrophages (BMDMs) were infected with three distinct Leishmania species (L. mexicana, L. major, L. infantum). Whole cell protein extracts from parasites and BMDMs were collected at seven different timepoints post-infection (ranging from 0.5 to 72 hours) and subjected to quantitative mass spectrometry analysis.
Our analysis enabled the quantification of approximately 2000 murine proteins across all three infection experiments. Specifically, 1500, 1000, and 1400 proteins were analyzed after infection with L. mexicana, L. major, and L. infantum, respectively. The number of differentially expressed proteins varied for each species over the time course of infection, with 349, 133, and 287 proteins identified for L. mexicana, L. major, and L. infantum, respectively.
Notably, our experiments uncovered numerous proteins potentially implicated in the infection process, which were not previously identified in transcriptome analyses. For instance, gene ontology enrichment revealed 41 L. mexicana protein IDs associated with oxidoreductase activity. Additionally, we identified 24 differentially expressed proteins lacking functional or structural annotation in the TriTryp database.
Currently, we are investigating several candidate proteins to elucidate novel mechanisms involved in parasite adaptation processes during macrophage infections.
Selected publications
The histone methyltransferase DOT1B is dispensable for stage differentiation and macrophage infection of Leishmania mexicana (2025) Eisenhuth N, Rauh ET, Mitnacht M, Debus A, Schleicher U, Butter F, Pruzinova K, Volf P, Janzen CJ. Front Cell Infect Microbiol. Jan 20;14:1502339. doi:10.3389/fcimb.2024.1502339.