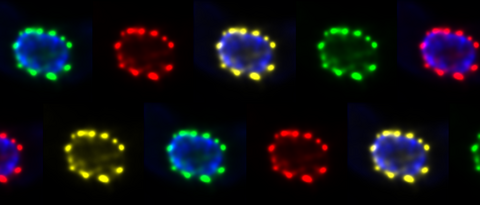
kramer lab
We study mRNA metabolism in early branched eukaryotes to understand aspects of eukaryogenesis.
Research synopsis
Recent publications
CURRENT LAB MEMBERS
NEWs
Dr. Paula Castaneda Londono

22nd September 2025
Many congratulations!
ALPH1 WEAVE project meeting before the BSP

1.9.-2.9.2025
Our WEAVE consortium met in České Budějovice to discuss our ongoing research on the development of drug candidates targeting ALPH1. We listened to the exciting research updates from our 1st year PhD students: Emmanuel, Alexia and Dawid. The Kramer, Zoltner and Gorna groups made working plans for the upcoming months. We also reported our work through conference talks and posters during the BSP Trypanosomiasis and Leishmaniasis Seminar (Sept 1-4. 2025).
Lab Pizza day. Do Nothing.

17 July 2025
Leticia and Alexia created the ultimate PhD game. We had a lot of fun in the beer garden after Bernardos defence.
Poster Presentation
5th November 2024
Johanna presents her fantastic data on streptavidin imaging in Caxambu.
Walter Colli Prize for Bernardo

7th of November, 2024
Bernardo wins the Walter Colli Prize at the Caxambu Meeting of the Brazilan Society for Protozoology for his talk about the trypanosome nuclear pore complex. Well done!
Leticia and Susanne in Lisna

10.-12.July 2024
Leticia and Susanne met with Martin Zoltner and the lab of Maria Gorna in Lisna (near Warsaw), for a phantastic lab-retreat (to remember!). Many thanks Maria, for a perfect organisation with the right priorities.
Sarah Monic is visiting

4th July 2024
Sarah Monic is visiting from Cambridge for an exciting talk on FISH TRYPANOSOMES.
ALPH meeting after the DDDS

15th March, 2024
We use the DDDS for a satellite meeting on ALPH1, to plan our next steps with our collaboraters from Warsaw and Prague. After intense discussions in the seminar room, diner in the Ratskeller was well deserved.
ALPH1 at the DDDS in Würzburg

12th to 15th of March, 2024
Leticia, Maria (3x), David and Susanne attend the DDDS meeting. ALPH1 is in focus: with 3 posters and one talk !
Bernardo and Johanna in Brazil 4

December, 2023
Johanna visits the laboratory of Angelea Cruz at the University of São Paulo. She also discussed her RNA Sequencing data with José Carlos Quilles Junior, who spent one year as a Postdoc in our lab in Würzburg.
Bernardo and Johanna in Brazil 3

December, 2023
Bernardo discusses science with the head of the FIOCRUZ institute, Dr. Stenio Perdigao Fragoso (picture), with Andrea Avila and many others.
Bernardo and Johanna in Brazil 2

5th of December, 2023
Johanna talks about her freshly submitted data, on how to use streptavidin to literally solve all antibody problems.
Bernardo and Johanna in Brazil 1

5th of December, 2023
Bernardo talks about his data on the organisation of the Trypanosome nuclear pore complex in his old institute, at FIOCRUZ Parana, in Curitiba.
Visitors from Brazil

11th-13th of September, 2023
Osvaldo Pompilio de Melo Neto from Instituto Aggeu Magalhães/Fundação Oswaldo Cruz and Fabiola Holetz from FIOCRUZ - PARANÁ
have been visiting the lab. Osvaldo gave a great talk about trypanosome translation initiation factors and we had phantastic scientific discussions, on projects of ongoing and future colaborations.
Nancy Standart visits for Bernardos PhD progress report

21-22th August 2022
Bernardo gave a wonderful talk. Thanks to all members of the thesis commitee and in particular to Nancy, for very helpful input and discussions.
Susanne in Brazil

Jun 2023
Susanne visits our long-term collaborator Fabiola Holetz at the Carlos Chagas Insitute in Curitiba.
Labmeeting in Prague

9th of February, 2023
Johanna und Susanne visit Prague, to discuss APEX and Co with Martin, SIqi und Farnaz.
спасибо GORBI

30th August 2022
... the Kramer lab would not exist without you.
RIP
Nuclear export in Potsdam

Jul 2022
Johanna and Bernardo present their work at the DGZ meeting "Life at the edge: the nuclear envelope and nucleocytoplasmic transport".
Visitors from Prague
11th to 13th May, 2022
Our collaborators from Martin Zoltners lab in Prague visited. We had a great and productive time together, discussing our joined project and plans for the next grant application.
Discussing ALPH in Warsaw

7th Dec 2021
Start up meeting in Warsaw with Maria GoRNA and Martin Zolter to plan how to employ ALPH as a drug target.
Joined Lab-Meeting in Prague

Sep 2021
We had our first in-person meeting since Corona: with our collaborators from the Zoltner lab at the Biocev near Prague. Zoom can never replace real meetings.
We have moved to our new lab!
2nd and 3rd June, 2021
This week we have moved into our new lab and we even found a place for the picture that we inherited. There are still some small issues to sort out, but in principle the lab is already fully functional and we look forwards to testing it with exciting experiments.
14 DAYS of Quarantine

April/May 2021
Covid once again, Lisa got the virus, but nobody else got infected :-). Here are images from our (almost) daily quarantine Zoom Meetings.
First PhD defence of the Kramer lab
14th April 2021
The Kramer lab has its first Doktor! Carina Goos successfully defended her Thesis. Many congratulations, Carina!
RNA 2019
June, 2019
We met with our Polish Collaboraters from the laboratory of Maria Gorna at the RNA society meeting in Krakow. Together we will unravel the secrets of the trypanosome decapping enzyme !
RESEARCH TOPICS
mRNA granules
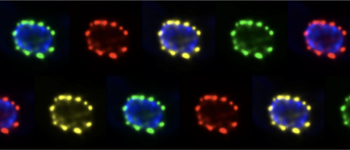
Many non-polysomal mRNAs aggregate into RNA granules, large ribonucleoprotein particles. What is the composition and function of RNA granules and how are they regulated?
mRNA decay
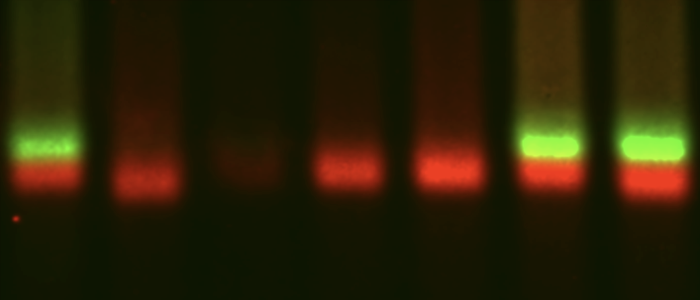
The first step in mRNA decay is the removal of the 5' cap. In trypanosomes, this is done by a highly unusual enzyme, an ApaH like phosphatase.
mRNA export

We recently found that trypanosomes can export their mRNAs co-transcriptionally.
PhD, Postdoc
Dr. Leticia Pereira (2025), Postdoc
Dr. Bernardo Papini Gabiatti (2025), PhD student
Do trypanosomes control nuclear export of mRNAs?
Dr. José Carlos Quilles Junior (2022-2023), Postdoc
ncRNAs in Leishmania
Paula Castaneda Londono (2022), PhD student
In vitro characterisation of the mRNA decapping enzyme ALPH1
Dr. Claudia Moreira (2019), Postdoc
Establishing novel proximity labelling techniques (Postdoc)
Dr. Carina Goos (2017 / Defense 2021), PhD student
Purification and characterisation of trypanosome nuclear periphery granules
Master Students
Luka-Michael Jekic (2025)
Lokalisierung und Funktion von Kernporenproteinen in Trypanosoma brucei
Benjamin Dietz (2025)
Lokalisation von möglichen Spindelpolkörper-Proteinen in Trypanosoma brucei
Carolin Schlitz (2024)
Analysis of mRNA decay pathways in Euglenozoa
Lilith Babilon/Ruis (2023)
Functional analysis of the mRNA decapping complex in Trypanosoma brucei
Johanna Odenwald (2021)
Interactions and Function of a novel mRNA decapping enzyme from Trypanosoma brucei
Andrea Reichert (2018)
Untersuchungen zur Funktion der spezifischen Lokalisation des T. brucei mRNA-Decapping-Enzyms
Nadja Sauer (2016)
In vivo Markierung von mRNAs in RNP Granula durch photoaktivierbare, abstandsabhängige Biotinylierung: Etablierung der Methode
Hanna Thoma (2016)
Warum sind für ribosomale Proteine kodierende mRNAs aus Stress-Granula ausgeschlossen?
Christina Julia Lorenz (2015, extern in Cambridge)
Identification of interaction partners of the DEAD-box RNA helicase DHH1 in Trypanosoma brucei
Nina Krienitz (2015, extern Cambridge and Dundee)
Implementation of a novel cryomilling method to analyse protein and RNA interactions of the poly(A)-binding proteins in Trypanosoma brucei
Melanie Fritz (2014)
Zusammensetzung und Ultrastruktur von Stress-Granula in Trypanosoma brucei brucei
Bachelor Students
Ann-Sophie Eisele (2025)
Establishing iterative Ultra Expansion in Trypanosoma brucei
Maya Wacker (2025)
Die Funktion des TREX2 Komplexes in Trypanosomen
Lea Sophie-Löffler (2024)
Can Euglena be used for Antibiotics discovery?
Amelie Eder (2022)
Untersuchungen zum Interaktom des Trypanosomen mRNA Decapping Enzyms ALPH1
Marko Korb (2022)
Serum-Abhängigkeit von Trypanosoma brucei brucei Kulturen: Untersuchungen zur Reduktion der benötigten FCS-Konzentration
Lisa Marie-Hofacker (2021)
Etablierung einer induzierbaren Bio-ID Methode in Trypanosoma brucei
Nicole Banholzer (2019)
Charakterisierung eines neuen mRNA Decapping-Enzyms aus Trypanosomen
Anna Sophie Kreis (2019)
Etablierung einer Methode zur mRNA-Visualisierung mittels CRISPR/dCAS13
Laura Gauglitz (2018)
Etablierung einer neuen BioID Methode in Trypanosomen
Laura Peters (2017)
Warum sind mRNAs, die für ribosomale Proteine kodieren, aus Stress-Granula ausgeschlossen?
Till Elharrar (2015)
Validierung der Lokalisation zu ‘Nuclear periphery granules’ von neu identifizierten Proteinkandidaten
Nadja Sauer (2014)
Überprüfung neuer Stressgranula-Protein-Kandidaten in Trypanosoma brucei
Daja Schichler (2014)
Analyse von RNA-Granula in Trypanosoma brucei
Melanie Fritz (2012)
Charakterisierung von Stress-Granula in Trypanosoma brucei brucei
Mario Hofweber (2012)
Ist das trypanosomale P-body Protein SCD6 ein Multimer?
Internship
Alae Benharoual (2024), Erasmus
Nadine Knoch (2024)
Lukas Herold (2023(24)
Niloofar Kalantari (2023)
Johanna Meyer (2023)
Nico Ankenbrand (2022)
Delara Shahidi (2021)
Sisco Jung (2021)
Frederik Heim
Corinna Kronenthaler
Johannes Schmidt
Bilal Tetik
Susheela Behera